Gaining Insights into Folding/Unfolding Protein Structures and their Importance for Several Applications: Historical Research Generated in the Biophysical Chemistry Area
DOI:
https://doi.org/10.29356/jmcs.v68i4.2297Keywords:
Protein stability, Folding and unfolding, Fluorescence and Circular Dichroism spectroscopy, Differential scanning calorimetry, KineticsAbstract
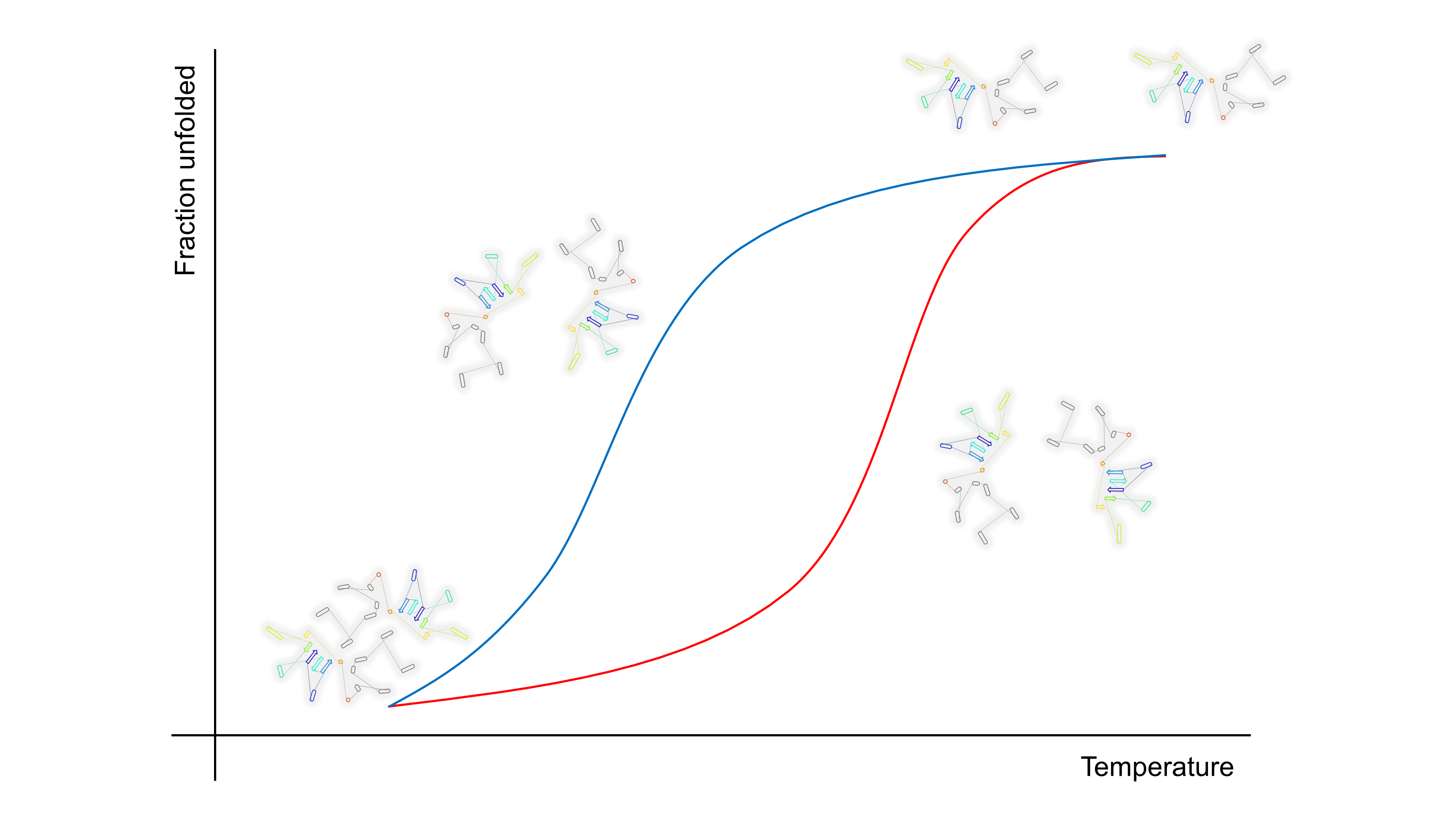
The research largely focuses on investigating the mechanisms of protein folding and unfolding in proteins, namely triosephosphate isomerase, glucosamine-6-phosphate deaminase, laccase, and bacteriophage M13. The article examines the mechanisms of protein denaturation and renaturation using kinetic equations, thermodynamic models, and molecular dynamics (MD) simulations. These results enhance our understanding of the thermodynamic and kinetic characteristics of these proteins. Furthermore, the study highlights the importance of conserved residues, as well as the influence of environmental conditions such as pH and temperature on protein stability and folding. These discoveries have potential implications in biotechnology and medicine, including the creation of protein-based products and therapies for infectious diseases, and neurodegenerative disorders. The paper acknowledges the groundbreaking contributions of Dr. Andrés Hernández Arana to the field of protein physical chemistry in México. His work has greatly influenced the progress of research in the areas of protein stability and kinetics.
Resumen. La investigación se centra en los mecanismos de plegado y desplegado de proteínas; estos mecanismos incluyen la triosafosfato isomerasa, la glucosamina-6-fosfato desaminasa, lacasa y el bacteriófago M13. Se utilizan ecuaciones cinéticas, modelos termodinámicos y simulaciones de dinámica molecular (MD) para analizar los mecanismos de desnaturalización y renaturalización de proteínas. Estos hallazgos nos ayudan a comprender mejor las características cinéticas y termodinámicas de estas proteínas. Además, el estudio destaca la importancia de los residuos conservados y puentes salinos en las proteínas, así como el impacto de los factores ambientales como el pH y la temperatura en la estabilidad y el plegado de las proteínas. Estos hallazgos tienen repercusiones en los campos de la biotecnología y la medicina, como la creación de productos y terapias basados en proteínas para enfermedades infecciosas y trastornos neurodegenerativos. El artículo reconoce el trabajo pionero del Dr. Andrés Hernández Arana en México en el campo de la termodinámica de proteínas. Su trabajo ha sido fundamental para el avance de la investigación en las áreas de cinética y estabilidad de proteínas.
Downloads
References
Galano-Frutos, J. J.; Nerín-Fonz, F.; Sancho, J. Journal of Chemical Information and Modeling. 2023, 63, 7791-7806. DOI: https://doi.org/10.1021/acs.jcim.3c01107.
Mei, G. Encyclopedia of Life Sciences. 2017, 1-7. DOI: https://doi.org/10.1002/9780470015902.A0027584.
Kelly, S. M.; Price, N. C. Encyclopedia of Life Sciences. 2009. DOI: https://doi.org/10.1002/9780470015902.A0003043.PUB2.
Wieczorek, G.; Niedzialek, D. Encyclopedia of Life Sciences. 2020, 1-18. DOI: https://doi.org/10.1002/9780470015902.A0003048.PUB3.
Johnson, C. M. Archives of Biochemistry and Biophysics. 2013, 531, 100-109. DOI: http://dx.doi.org/10.1016/j.abb.2012.09.008. DOI: https://doi.org/10.1016/j.abb.2012.09.008
Sanchez-Ruiz, J. M. Subcellular Biochemistry 1995. DOI: https://doi.org/10.1007/978-1-4899-1727-0_6.
Hernbndez-Arana, A.; Rojo-Dominguez, A.; Altamirano, M. M.; Calcagno, M. L. Differential Scanning Calorimetry of the Irreversible Denaturation of Escherichia coli Glucosamine-6-phosphate Deaminase?; 1993. DOI: https://doi.org/10.1021/bi00065a017
Kuril, A. K. Journal of Pharmaceutical Research International. 2024, 36, 179-187. DOI: https://doi.org/10.9734/jpri/2024/v36i77549.
Nakama, T.; Rossen, A.; Ebihara, R.; Yagi-Utsumi, M.; Fujita, D.; Kato, K.; Sato, S.; Fujita, M. Chemical Science 2023, 14, 2910-2914. DOI: https://doi.org/10.1039/D2SC05879K.
Barrett, J. The International Journal of Biochemistry & Cell Biology. 2001, 33 2, 105-117. DOI: https://doi.org/10.1016/S1357-2725(00)00083-2.
Durowoju, I. B.; Bhandal, K. S.; Jian Hu, B. C.; Kirkitadze, M. Journal of Visualized Experiments. 2017, 121 (e55262). DOI: https://doi.org/10.3791/5526.
Greenfield, N. J. Nature Protocols 2007, 1, 2527-2535. DOI: https://doi.org/10.1038/nprot.2006.204.
Preeti Gupta, A. I.; Ahmad, F.; Hassan, M. I. Protein Folding Dynamics and Stability. 2023. DOI: https://doi.org/10.1007/978-981-99-2079-2_1.
Seelig, J.; Schönfeld, H.-J. Thermal protein unfolding by differential scanning calorimetry and circular dichroism spectroscopy Two-state model versus sequential unfolding. Quarterly Reviews of Biophysics 2016, 49, e9. From Cambridge University Press Cambridge Core. DOI: https://doi.org/10.1017/S0033583516000044.
Lakowicz, J. R. 10. Protein Fluorescence BT - Principles of Fluorescence Spectroscopy. 2006, 1-47. DOI: https://doi.org/10.1007/978-0-387-46312-4_16.
dos Santos Rodrigues, F. H.; Delgado, G. G.; Santana da Costa, T.; Tasic, L. BBA Advances. 2023, 3. DOI: https://doi.org/10.1016/j.bbadva.2023.100091.
Michalet, X.; Weiss, S.; Jäger, M. Single-molecule fluorescence studies of protein folding and conformational dynamics. In Chemical Reviews, 2006; Vol. 106, pp 1785-1813. DOI: https://doi.org/10.1021/cr0404343
Basak, S.; Chattopadhyay, K. Studies of protein folding and dynamics using single molecule fluorescence spectroscopy. In Physical Chemistry Chemical Physics, Royal Society of Chemistry: 2014; Vol. 16, pp 11139-11149. DOI: https://doi.org/10.1039/c3cp55219e
Gooran, N.; Kopra, K. Fluorescence-Based Protein Stability Monitoring—A Review. In International Journal of Molecular Sciences, 2024; Vol. 25. DOI: https://doi.org/10.3390/ijms25031764
Yu, M.; Si, W.; Sha, J. Molecular Dynamics Simulation for Protein Unfolding. In 15th IEEE International Conference on Nano/Micro Engineered and Molecular System, NEMS 2020, 2020/9//, 2020; Institute of Electrical and Electronics Engineers Inc.: pp 382-386. DOI: https://doi.org/10.1109/NEMS50311.2020.9265552.
Caflisch, A.; Paci, E. Protein Folding Handbook. 2008, 2, 1143-1169. DOI: https://doi.org/10.1002/9783527619498.CH32. DOI: https://doi.org/10.1002/9783527610754.sf11
Scheraga, H. A.; Khalili, M.; Liwo, A. Protein-folding dynamics: Overview of molecular simulation techniques. In Annual Review of Physical Chemistry, Annual Reviews Inc.: 2007; Vol. 58, pp 57-83. DOI: https://doi.org/10.1146/annurev.physchem.58.032806.104614
Singh, Y.; Hocky, G. M. Improved Prediction of Molecular Response to Pulling by Combining Force Tempering with Replica Exchange Methods. The Journal of Physical Chemistry B. 2024, 128, 706-715. DOI: https://doi.org/10.1021/acs.jpcb.3c07081.
Kříž, P.; Beránek, J.; Spiwok, V. The Journal of Chemical Physics. 2024, 160, 184116. DOI: https://doi.org/10.1063/5.0204992 (acccessed 7/14/2024).
Benítez-Cardoza, C. G.; Rojo-Domínguez, A.; Hernández-Arana, A. Biochemistry. 2001, 40, 9049-9058. DOI: https://doi.org/10.1021/bi010528w.
González-Mondragón, E.; Zubillaga, R. A.; Saavedra, E.; Chánez-Cárdenas, M. E.; Pérez-Montfort, R.; Hernández-Arana, A. Biochemistry. 2004, 43 (11), 3255-3263. DOI: https://doi.org/10.1021/bi036077s.
Reyes-López, C. A.; González-Mondragón, E.; Benítez-Cardoza, C. G.; Chánez-Cárdenas, M. E.; Cabrera, N.; Pérez-Montfort, R.; Hernández-Arana, A. Proteins: Structure, Function and Genetics. 2008, 72, 972-979. DOI: https://doi.org/10.1002/prot.21994.
Cruces-Angeles, M. E.; Cabrera, N.; Perez-Montfort, R.; Reyes-Lopez, C. A.; Hernandez-Arana, A. Protein & Peptide Letters 2011, 18, 1290-1298. DOI: https://doi.org/10.2174/092986611797642715.
Toledo-Núñez, C.; López-Cruz, J. I.; Hernández-Arana, A. Biophysical Chemistry. 2012, 167, 36-42. DOI: https://doi.org/10.1016/j.bpc.2012.04.004.
Toledo-Núñez, C.; Vera-Robles, L. I.; Arroyo-Maya, I. J.; Hernández-Arana, A. Analytical Biochemistry. 2016, 509, 104-110. DOI: https://doi.org/10.1016/j.ab.2016.07.006.
González-Cansino, J. L.; Vieyra-Eusebio, M. T.; Vera-Robles, L. I.; Hernández-Arana, A. Thermochimica Acta. 2019, 672, 53-59. DOI: https://doi.org/10.1016/j.tca.2018.12.010.
García-Gutiérrez, P.; Camarillo-Cadena, M.; Vera-Robles, L. I.; Zubillaga, R. A.; Hernández-Arana, A. Spectrochimica Acta - Part A: Molecular and Biomolecular Spectroscopy 2022, 274. DOI: https://doi.org/10.1016/j.saa.2022.121039.
Hernández-Arana, A. Advances in Protein Physical Chemistry 2008, 139-154.
Ribeiro, A. J. M.; Holliday, G. L.; Furnham, N.; Tyzack, J. D.; Ferris, K.; Thornton, J. M. Nucleic Acids Research 2018, 46 (D1), D618-D623. DOI: https://doi.org/10.1093/NAR/GKX1012.
Junghare, V.; Bhattacharya, S.; Ansari, K.; Hazra, S. Markov State Models of Molecular Simulations to Study Protein Folding and Dynamics. In Protein Folding Dynamics and Stability: Experimental and Computational Methods, Saudagar, P., Tripathi, T. Eds.; Springer Nature Singapore, 2023; pp 147-164. DOI: https://doi.org/10.1007/978-981-99-2079-2_8
Arregui, L.; Ayala, M.; Gomez-Gil, X.; Gutierrez-Soto, G.; Hernandez-Luna, C. E.; Herrera de Los Santos, M.; Levin, L.; Rojo-Dominguez, A.; Romero-Martinez, D.; Saparrat, M. C. N.; et al. Laccases: structure, function, and potential application in water bioremediation. Microb Cell Fact 2019, 18 (1), 200. From NLM Medline. DOI: https://doi.org/10.1186/s12934-019-1248-0.

Downloads
Published
Issue
Section
License
Copyright (c) 2024 Leonardo David Herrera-Zuñiga, Arturo Rojo Domínguez

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.