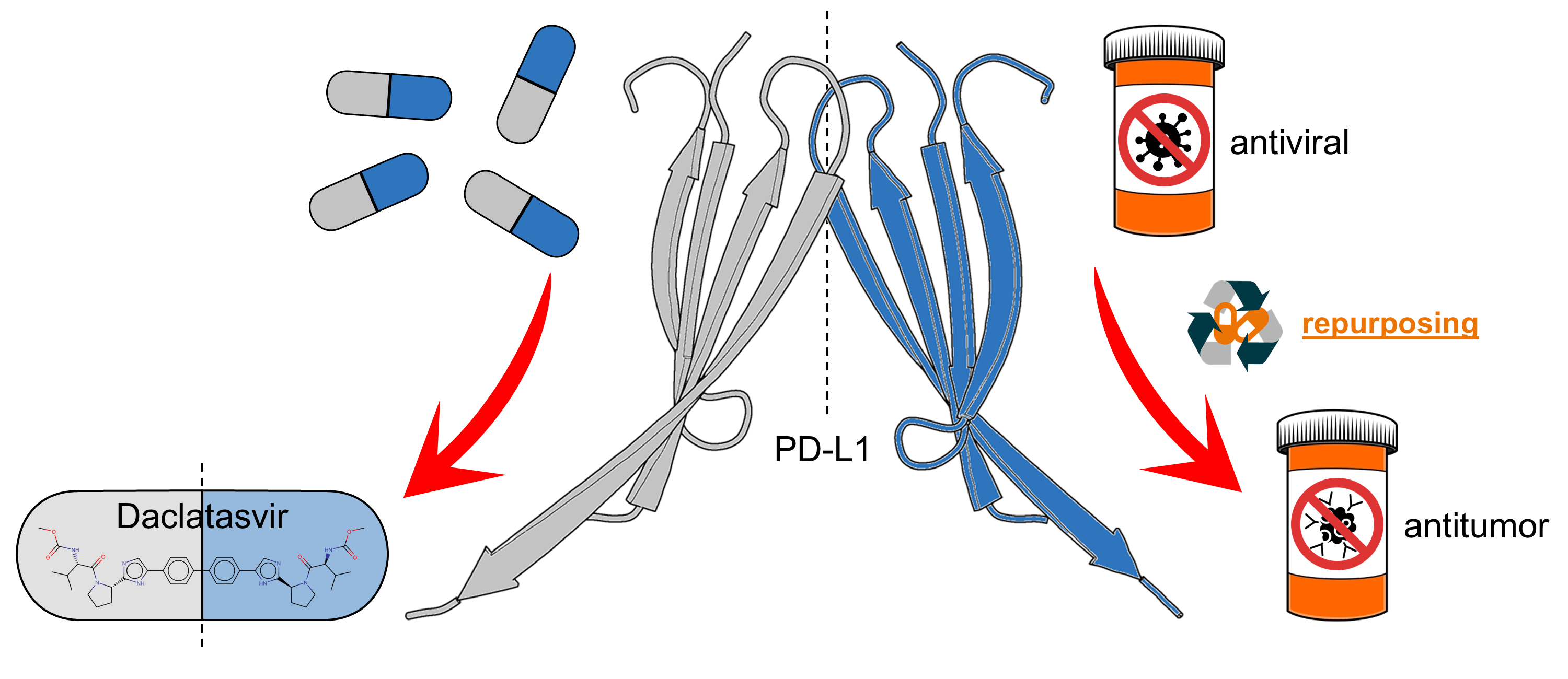
Daclatasvir, A Symmetric Drug for an Anti-Symmetric Target
DOI:
https://doi.org/10.29356/jmcs.v70i1.2393Keywords:
PD-L1, Daclatasvir, drug repurposing, virtual screening, cancer immunotherapyAbstract
Abstract. Immunotherapy has become a cornerstone in cancer treatment, with anti-PD-L1 antibodies effectively used across various cancers. Although these therapies have shown success, antibodies face limitations in bioavailability compared to low molecular mass compounds. An alternative strategy is to stabilize PD-L1 homodimers to prevent their immunosuppressive activity. The homodimer interface forms a tunnel-like cavity that can accommodate small molecules. However, no small drugs targeting PD-L1 homodimers have been approved for cancer treatment. Drug repurposing offers a promising approach to bridge this gap. In this study, we sought to identify potential PD-L1 inhibitors among FDA-approved drugs using virtual screening, followed by molecular docking, molecular dynamics simulations, and MM/PBSA binding energy calculations. Our results indicate that daclatasvir, an FDA-approved antiviral for hepatitis C, forms a stable and energetically favorable complex with the PD-L1 homodimer, suggesting it as a promising candidate for further investigation in cancer immunotherapy. Due to its symmetry, daclatasvir simultaneously interacts with both PD-L1 monomers in an equivalent manner, bridging the dimer interface. Its biphenyl core anchors at the center of the tunnel, the imidazole rings position at the entrances, and the pyrrolidine rings remain exposed to the solvent. Our in-depth characterization of the binding mode of daclatasvir clarifies its binding mechanism, and recent experimental findings have also indicated that daclatasvir binds to PD-L1, supporting its potential in this new context.
Resumen. La inmunoterapia se ha convertido en una piedra angular en el tratamiento del cáncer, y los anticuerpos anti-PD-L1 se utilizan eficazmente en varios tipos de cáncer. Aunque estas terapias han demostrado ser exitosas, los anticuerpos enfrentan limitaciones de biodisponibilidad en comparación con los compuestos de baja masa molecular. Una alternativa al uso de anticuerpos consiste en estabilizar homodímeros de PD-L1 para impedir su función inmunosupresora. La interfase de los homodímeros de PD-L1 constituye un túnel que puede alojar moléculas de baja masa molecular. Sin embargo, no existen moléculas pequeñas dirigidas al homodímero de PD-L1 con aprobación regulatoria para el tratamiento del cáncer. El reposicionamiento de fármacos ofrece un enfoque prometedor para cerrar esta brecha. En este estudio, buscamos identificar potenciales inhibidores de PD-L1 entre los fármacos aprobados por la FDA mediante cribado virtual, acoplamientos moleculares, simulaciones de dinámica molecular y cálculos de energía de unión MM/PBSA. Nuestros resultados indican que daclatasvir, un antiviral aprobado por la FDA para la hepatitis C, forma un complejo estable y energéticamente favorable con el homodímero PD-L1, lo que sugiere que es un candidato prometedor en la inmunoterapia contra el cáncer. Debido a su simetría, daclatasvir puede interactuar simultáneamente y de la misma manera con ambos monómeros de PD-L1, estabilizando la unión. El núcleo bifenilo de daclatasvir se aloja en el centro del túnel del homodímero, los anillos de imidazol se colocan en las entradas, y los anillos de pirrolidina permanecen expuestos al solvente. Nuestra caracterización detallada del modo de unión de daclatasvir aclara su mecanismo de interacción. Además, hallazgos experimentales recientes indican que daclatasvir se une a PD-L1, lo que respalda su potencial en este nuevo contexto.
Downloads
References
1. Zhang, Y.; Zheng, J., in: Regulation of Cancer Immune Checkpoints: Molecular and Cellular Mechanisms and Therapy; Xu, J., Ed.; Springer: Singapore, 2020, 201–226. DOI: https://doi.org/10.1007/978-981-15-3266-5_9
2. Zak, K. M.; Grudnik, P.; Magiera, K.; Dömling, A.; Dubin, G.; Holak, T. A. Structure. 2017, 25, 1163–1174. DOI: https://doi.org/10.1016/j.str.2017.06.011
3. Hui, E.; Cheung, J.; Zhu, J.; Su, X.; Taylor, M. J.; Wallweber, H. A.; Sasmal, D. K.; Huang, J.; Kim, J. M.; Mellman, I.; Vale, R. D. Science. 2017, 355, 1428–1433. DOI: https://doi.org/10.1126/science.aaf1292
4. Marcucci, F.; Rumio, C.; Corti, A. Biochim. Biophys. Acta Rev. Cancer. 2017, 1868, 571–583. DOI: https://doi.org/10.1016/j.bbcan.2017.10.006
5. Jiang, X.; Wang, J.; Deng, X.; Xiong, F.; Ge, J.; Xiang, B.; Wu, X.; Ma, J.; Zhou, M.; Li, X.; Li, Y.; Li, G.; Xiong, W.; Guo, C.; Zeng, Z. Mol. Cancer. 2019, 18, 10.DOI: https://doi.org/10.1186/s12943-018-0928-4
6. Patel, S. P.; Kurzrock, R. Mol. Cancer Ther. 2015, 14, 847–856. DOI: https://doi.org/10.1158/1535-7163.MCT-14-0983
7. Ullah, A.; Pulliam, S.; Karki, N. R.; Khan, J.; Jogezai, S.; Sultan, S.; Muhammad, L.; Khan, M.; Jamil, N.; Waheed, A.; Belakhlef, S.; Ghleilib, I.; Vail, E.; Heneidi, S.; Karim, N. A. Clin. Prac. 2022, 12, 653–671. DOI: https://doi.org/10.3390/clinpract12050068
8. Wang, X.; Teng, F.; Kong, L.; Yu, J. OncoTargets Ther. 2016, 9, 5023–5039. DOI: https://doi.org/10.2147/OTT.S105862
9. Jiang, M.; Liu, M.; Liu, G.; Ma, J.; Zhang, L.; Wang, S. mAbs. 2023, 15, 2236740. DOI: https://doi.org/10.1080/19420862.2023.2236740
10. Esfahani, K.; Roudaia, L.; Buhlaiga, N.; Del Rincon, S. V.; Papneja, N.; Miller, W. H. A Curr. Oncol. 2020, 27, S87–S97. DOI: https://doi.org/10.3747/co.27.5223
11. Farkona, S.; Diamandis, E. P.; Blasutig, I. M. BMC Med. 2016, 14, 73. DOI: https://doi.org/10.1186/s12916-016-0623-5
12. Naidoo, J.; Page, D. B.; Li, B. T.; Connell, L. C.; Schindler, K.; Lacouture, M. E.; Postow, M. A.; Wolchok, J. D. Ann. Oncol. 2015, 26, 2375–2391. DOI: https://doi.org/10.1093/annonc/mdv383
13. Zhan, M.-M.; Hu, X.-Q.; Liu, X.-X.; Ruan, B.-F.; Xu, J.; Liao, C. Drug Discov. Today. 2016, 21, 1027–1036. DOI: https://doi.org/10.1016/j.drudis.2016.04.011
14. Liu, G.; Kang, S.; Wang, X.; Shang, F. Front. Oncol. 2021, 11. DOI: https://doi.org/10.3389/fonc.2021.669195
15. Hu, X.; Hay, J. W. Lung Cancer. 2018, 123, 166–171. DOI: https://doi.org/10.1016/j.lungcan.2018.07.012
16. Bailly, C.; Vergoten, G. Biochem. Pharmacol. 2020, 174, 113821. DOI: https://doi.org/10.1016/j.bcp.2020.113821
17. Liu, L.; Zhang, H.; Hou, J.; Zhang, Y.; Wang, L.; Wang, S.; Yao, Z.; Xie, T.; Wen, X.; Xu, Q.; Dai, L.; Feng, Z.; Zhang, P.; Wu, Y.; Sun, H.; Liu, J.; Yuan, H. J. Med. Chem. 2024, 67, 4977–4997. DOI: https://doi.org/10.1021/acs.jmedchem.4c00102
18. Cheng, B.; Wang, W.; Liu, T.; Cao, H.; Pan, W.; Xiao, Y.; Liu, S.; Chen, J. Signal Transduction Target Ther. 2023, 8, 91. DOI: https://doi.org/10.1038/s41392-022-01292-5
19. Gashaw, I.; Ellinghaus, P.; Sommer, A.; Asadullah, K. Drug Discov. Today. 2011, 16, 1037–1043. DOI: https://doi.org/10.1016/j.drudis.2011.09.007
20. Makley, L. N.; Gestwicki, J. E. Chem. Biol. Drug Des. 2013, 81, 22–32. DOI: https://doi.org/10.1111/cbdd.12066
21. Berman, H. M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T. N.; Weissig, H.; Shindyalov, I. N.; Bourne, P. E. Nucleic Acids Res. 2000, 28, 235–242. DOI: https://doi.org/10.1093/nar/28.1.235
22. Klimek, J.; Kruc, O.; Ceklarz, J.; Kamińska, B.; Musielak, B.; van der Straat, R.; Dömling, A.; Holak, T. A.; Muszak, D.; Kalinowska-Tłuścik, J.; Skalniak, Ł.; Surmiak, E. Molecules. 2024, 29, 2646. DOI: https://doi.org/10.3390/molecules29112646
23. Chen, R.; Yuan, D.; Ma, J. Future Med. Chem. 2022, 14, 97–113. DOI: https://doi.org/10.4155/fmc-2021-0256
24. Basu, S.; Yang, J.; Xu, B.; Magiera-Mularz, K.; Skalniak, L.; Musielak, B.; Kholodovych, V.; Holak, T. A.; Hu, L. J. Med. Chem. 2019, 62, 7250–7263. DOI: https://doi.org/10.1021/acs.jmedchem.9b00795
25. Kawashita, S.; Aoyagi, K.; Yamanaka, H.; Hantani, R.; Naruoka, S.; Tanimoto, A.; Hori, Y.; Toyonaga, Y.; Fukushima, K.; Miyazaki, S.; Hantani, Y. Bioorg. Med. Chem. Lett. 2019, 29, 2464–2467. DOI: https://doi.org/10.1016/j.bmcl.2019.07.027
26. Pushpakom, S.; Iorio, F.; Eyers, P. A.; Escott, K. J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; Norris, A.; Sanseau, P.; Cavalla, D.; Pirmohamed, M. Nat. Rev. Drug Discov. 2019, 18, 41–58. DOI: https://doi.org/10.1038/nrd.2018.168
27. Jourdan, J.-P.; Bureau, R.; Rochais, C.; Dallemagne, P. J. Pharm. Pharmacol. 2020, 72, 1145–1151. DOI: https://doi.org/10.1111/jphp.13273
28. Bellera, C. L.; Di Ianni, M. E.; Sbaraglini, M. L.; Castro, E. A.; Bruno-Blanch, L. E.; Talevi, A. in: Frontiers in Computational Chemistry; Ul-Haq, Z., Madura, J. D., Eds.; Bentham Science Publishers, 2015, 44–81. DOI: 10.2174/9781608058648115010004.
29. Cheng, F.; Desai, R. J.; Handy, D. E.; Wang, R.; Schneeweiss, S.; Barabási, A.-L.; Loscalzo, J. Nat. Commun. 2018, 9, 2691. DOI: https://doi.org/10.1038/s41467-018-05116-5
30. Mullins, J. G. L. Biochem. Soc. Trans. 2022, 50, 747–758. DOI: https://doi.org/10.1042/BST20200967
31. Hodos, R. A.; Kidd, B. A.; Shameer, K.; Readhead, B. P.; Dudley, J. T. WIREs Syst. Biol. Med. 2016, 8, 186–210. DOI: https://doi.org/10.1002/wsbm.1337
32. Kumar, S.; Kumar, S. in: In Silico Drug Design; Roy, K., Ed.; Academic Press, 2019, 161–189. DOI: https://doi.org/10.1016/B978-0-12-816125-8.00006-7
33. Sohraby, F.; Bagheri, M.; Aryapour, H., in: Performing an In Silico Repurposing of Existing Drugs by Combining Virtual Screening and Molecular Dynamics Simulation. In Computational Methods for Drug Repurposing; Vanhaelen, Q., Ed.; Springer: New York, NY, 2019, 23–43. DOI: https://doi.org/10.1007/978-1-4939-8955-3_2
34. McConachie, S. M.; Wilhelm, S. M.; Kale-Pradhan, P. B. Expert Rev. Clin. Pharmacol. 2016, 9, 287–302. DOI: https://doi.org/10.1586/17512433.2016.1129272
35. Kumari, R.; Kumar, R.; Lynn, A. J. Chem. Inf. Model. 2014, 54, 1951–1962. DOI: https://doi.org/10.1021/ci500020m
36. Sun, M.; Lv, S.; Pan, Y.; Song, Q.; Ma, C.; Yu, M.; Gao, X.; Guo, X.; Wang, S.; Gao, Z.; Wang, S.; Meng, Q.; Zhang, L.; Li, Y. Bioorg. Chem. 2024, 153, 107874. DOI: https://doi.org/10.1016/j.bioorg.2024.107874
37. Kim, S. Curr. Protoc. 2021, 1, e217. DOI: https://doi.org/10.1002/cpz1.217
38. Knox, C.; Wilson, M.; Klinger, C. M.; Franklin, M.; Oler, E.; Wilson, A.; Pon, A.; Cox, J.; Chin, N. E. Strawbridge, S. A.; Garcia-Patino, M.; Kruger, R.; Sivakumaran, A.; Sanford, S.; Doshi, R.; Khetarpal, N.; Fatokun, O.; Doucet, D.; Zubkowski, A.; Rayat, D. Y.; Jackson, H.; Harford, K.; Anjum, A.; Zakir, M.; Wang, F.; Tian, S.; Lee, B.; Liigand, J.; Peters, H.; Wang, R. Q.; Nguyen, T.; So, D.; Sharp, M.; da Silva, R.; Gabriel, C.; Scantlebury, J.; Jasinski, M.; Ackerman, D.; Jewison, T.; Sajed, T.; Gautam, V.; Wishart, D. S. Nucleic Acids Res. 2024, 52, D1265–D1275. DOI: https://doi.org/10.1093/nar/gkad976
39. Bogdanov, A.; Bogdanov, A.; Chubenko, V.; Volkov, N.; Moiseenko, F.; Moiseyenko, V. Front. Oncol. 2022, 12. DOI: https://doi.org/10.3389/fonc.2022.979154
40. o, S.; Cheng, X.; Lee, J.; Kim, S.; Park, S.-J.; Patel, D. S.; Beaven, A. H.; Lee, K. I.; Rui, H.; Park, S.; Lee, H. S.; Roux, B.; MacKerell, A. D.; Klauda, J. B.; Qi, Y.; Im, W. J. Comput. Chem. 2017, 38, 1114–1124. DOI: https://doi.org/10.1002/jcc.24660
41. Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B. L.; Grubmüller, H.; MacKerell, A. D. Nat. Methods. 2017, 14, 71–73. DOI: https://doi.org/10.1038/nmeth.4067
42. Abraham, M. J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J. C.; Hess, B.; Lindahl, E. SoftwareX. 2015, 1–2, 19–25. DOI: https://doi.org/10.1016/j.softx.2015.06.001
43. Genheden, S.; Ryde, U. Expert Opin. Drug Discov. 2015, 10, 449–461. DOI: https://doi.org/10.1517/17460441.2015.1032936
44. Saad, K. A.; Eldawy, M. A.; Elokely, K. M. J. Mol. Model. 2022, 29, 25. DOI: https://doi.org/10.1007/s00894-022-05420-4
45. Langhans, B.; Nischalke, H. D.; Krämer, B.; Hausen, A.; Dold, L.; van Heteren, P.; Hüneburg, R.; Nattermann, J.; Strassburg, C. P.; Spengler, U. J. Hepatol. 2017, 66, 888–896. DOI: https://doi.org/10.1016/j.jhep.2016.12.019
46. Guo, J.; Yu, F.; Zhang, K.; Jiang, S.; Zhang, X.; Wang, T. RSC Med. Chem. 2024, 15, 1096–1108. DOI: https://doi.org/10.1039/D3MD00636K
47. Wang, T.; Cai, S.; Cheng, Y.; Zhang, W.; Wang, M.; Sun, H.; Guo, B.; Li, Z.; Xiao, Y.; Jiang, S. J. Med. Chem. 2022, 65, 3879–3893. DOI: https://doi.org/10.1021/acs.jmedchem.1c01682
48. Szereday, L.; Meggyes, M.; Berki, T.; Miseta, A.; Farkas, N.; Gervain, J.; Par, A.; Par, G. Clin. Exp. Med. 2020, 20, 219–230. DOI: https://doi.org/10.1007/s10238-020-00618-3
49. Ganesan, A.; Ahmed, M.; Okoye, I.; Arutyunova, E.; Babu, D.; Turnbull, W. L.; Kundu, J. K.; Shields, J.; Agopsowicz, K. C.; Xu, L.; Tabana, Y.; Srivastava, N.; Zhang, G.; Moon, T. C.; Belovodskiy, A.; Hena, M.; Kandadai, A. S.; Hosseini, S. N.; Hitt, M.; Walker, J.; Smylie, M.; West, F. G.; Siraki, A. G.; Lemieux, M. J.; Elahi, S.; Nieman, J. A.; Tyrrell, D. L.; Houghton, M.; Barakat, K. Sci. Rep. 2019, 9, 12392. DOI: https://doi.org/10.1038/s41598-019-48826-6
50. Liu, M.; Zhang, Y.; Guo, Y.; Gao, J.; Huang, W.; Dong, X. Med. Chem. Res. 2021, 30, 1230–1239. DOI: https://doi.org/10.1007/s00044-021-02728-3
51. Jafari, M.; Guan, Y.; Wedge, D. C.; Ansari-Pour, N. Genome Biol. 2021, 22, 71. DOI: https://doi.org/10.1186/s13059-021-02292-4
52. Pillai, M.; Wu, D. AMIA Annu. Symp. Proc. 2023, 559. DOI: https://doi.org/10.17615/dmfd-x198
53. Nat. Comput. Sci. 2023, 3, 361–361. DOI: https://doi.org/10.1038/s43588-023-00462-x

Downloads
Published
Issue
Section
License
Copyright (c) 2025 Luis Cordova-Bahena, Axel Sánchez-Álvarez, Ivonne López-Lerma, Nohemí Salinas-Jazmín, José L. Medina-Franco, Marco Velasco-Velázquez

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.