Targeted transcriptomics of the Mexican prickly poppy (Argemone mexicana L.) reveals diverse proteins related to benzylisoquinoline alkaloid biosynthesis
DOI:
https://doi.org/10.29356/jmcs.v69i4.2223Keywords:
Argemone mexicana, benzylisoquinoline alkaloids, cytochrome P450, dihydrobenzophenantridine oxidase, sanguinarine reductase, tetrahydroprotoberberine oxidase, transcriptomicsAbstract
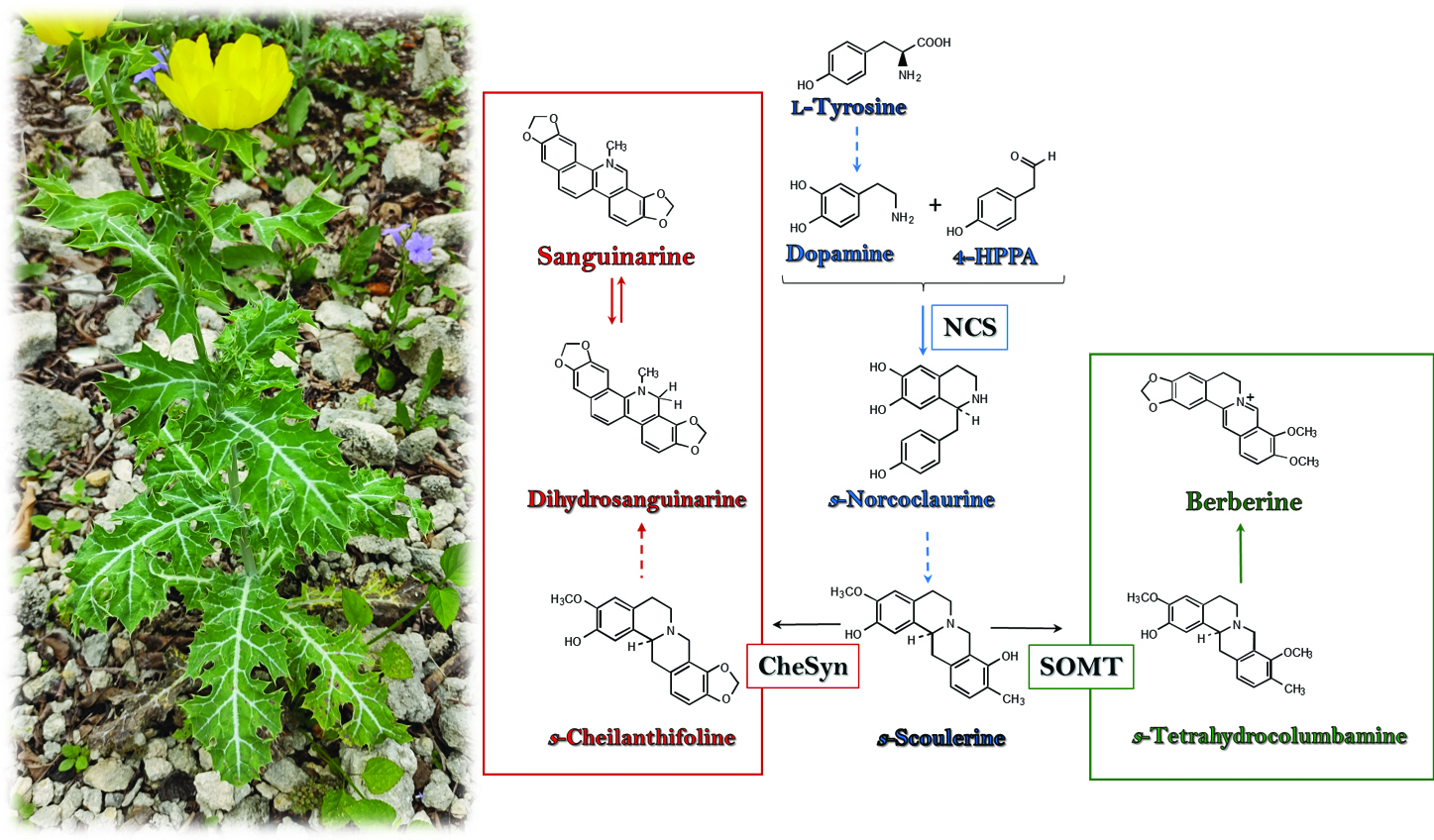
Abstract. A transcriptomic approach was employed to describe a set of putatively protein-coding sequences involved in the biosynthesis of berberine and sanguinarine, the two major benzylisoquinoline alkaloids (BIA) from Argemone mexicana (L.; Papaveraceae). A robust de novo assembled transcriptome was obtained from developing seedlings. Initial screening identified 514 unigenes, from eight different Pfam domains, such as Cyt-P450 dependent proteins, which are recurrently involved in BIA biosynthesis. Additional annotation by KEGG Orthology and Gene Ontology supported putative participation of the selected proteins in alkaloid biosynthesis. Moreover, in silico structure prediction of sanguinarine reductase (SanR), dihydrobenzophenantridine oxidase (DHBO) and tetrahydroprotoberberine oxidase (STOX), involved in the last reactions of sanguinarine and berberine biosynthesis, fitted to those of previously characterized proteins from related species, and thus, further supporting proper annotation. Hence, the pipeline analysis presented can provide a comprehensive description of the biosynthetic potential of this plant through functionality associated to its transcripts.
Resumen. Un acercamiento transcriptómico se empleó para predecir un conjunto de secuencias condificantes para proteínas presuntamente involucradas en la biosíntesis de dos de los principales alcaloides bencilisoquinlínicos (ABI) en Argemone mexicana (L.; Papaveraceae). Un transcriptoma robusto, se obtuvo de plántulas en desarrollo. Un cribaje inicial identificó 514 unigenes de ocho dominios Pfam diferentes, incluyendo proteínas dependientes del CitP450, que participan en muchas reacciones en la biosíntesis de ABI. Anotaciones adicionales siguiendo la ortología KEGG y Gene Ontology sugirió la participación de un grupo de proteínas en la biosíntesis de alcaloides. Más aún, el modelaje estructural in silico de la sanguinarina reductasa (SanR), dihidrobenzophenantridina oxidasa (DHBO) y tetrahidroprotoberberine oxidase (STOX), responsables de las últimas reacciones de la síntesis de sanguinarina y berberina encajaron los previamente descritos para estas enzimas en otras especies relacionadas, confirmando la asignación recibida en la anotación. De este modo, el análisis bioinformático realizado puede ser útil para la descripción detallada del potencial biosintético de esta planta a través de la caracterización funcional de los candidatos seleccionados.
Downloads
References
1. Gbesso, G. H. F.; Gbesso, F. K.; Adoukonou, R. C. F.; Akabassi, G. C.; Padonou, E. A.; Tente, A. B. H. Ethnobot. Res. Appl. 2021, 21, 1–11. DOI: https://doi.org/10.32859/era.21.20.1-11.
2. Priya, C. L.; Rao, Kokati V. B. Int. J. Pharm. Sci. Res. 2012, 36, 2143–48. DOI: http://dx.doi.org/10.13040/IJPSR.0975-8232.3(7).2143-48
3. Rubio-Pina, J.; Vazquez-Flota, F. Curr. Top. Med. Chem. 2013, 13, 2200–2207. DOI: https://doi.org/10.2174/15680266113139990152.
4. Babu, C. K.; Khanna, S. K.; Das, M. Antioxid. Redox Signaling. 2007, 9, 515–525. DOI: https://doi.org/10.1089/ars.2006.1492.
5. Vázquez-Flota, F; Rubio-Piña, J.; Xool-Tamayo, J.; Vergara-Olivares, M.; Tamayo-Ordoñez, Y.; Monforte-González, M.; Guízar-González, C.; Mirón-López, G. Rev. Fitotec. Mex. 2018, 41, 13–21. DOI: https://doi.org/10.35196/rfm.2018.1.13-21.
6. Laines-Hidalgo, J. I.; Muñoz-Sánchez, J. A.; Loza-Müller, L.; Vázquez-Flota, F. Molecules, 2022, 27, 1378. DOI: https://doi.org/10.3390/molecules27041378.
7. Chang, Y. C.; Chang, F. R.; Khalil, A. T.; Hsieh, P. W.; Wu, Y. C. Zeitschrift für Naturforschung C. 2003, 58, 521–526. DOI: https://doi.org/10.1515/znc-2003-7-813.
8. Gali, K.; Ramakrishnan, G.; Kothai, R.; Jaykar, B. Int. J. Pharmtech. Res. 2011, 3,1329–1333.
9. Nayak, P.; Kar, D. M.; Maharana, L. Pharmacologyonline. 2011, 1, 889–903.
10. Zhang, B. Y.; Chen, M.; Chen, X. C.; Cao, K.; You, Y.; Qian, Y. J.; Yu, W. K. Br. J. Surg. 2021, 108, e9–e11. DOI: https://doi.org/10.1093/bjs/znaa021.
11. Namdeo, A. G.; Jadhav, T. A.; Rai, P. K.; Gavali, S.; Mahadik, K. Phcog. Rev. 2007, 1, 227–231.
12. Isah, T.; Umar, S.; Mujib, A.; Sharma, M. P.; Rajasekharan, P. E.; Zafar, N.; Frukh, A. Plant Cell Tiss. Organ Cult. 2018, 132, 239–265. DOI: https://doi.org/10.1007/s11240-017-1332-2.
13. Yadav, A. N.; Kour, D.; Rana, K. L.; Yadav, N.; Singh, B.; Chauhan, V. S.; Rastegari, A. A.; Hesham, A. E.; Gupta, V. K., in: New and Future Developments in Microbial Biotechnology and Bioengineering: Microbial Secondary Metabolites Biochemistry and Applications. Chapter 20, Vijai Kumar Gupta and Anita Pandey, Elsevier, 2019, 279-320. DOI: https://doi.org/10.1016/B978-0-444-63504-4.00020-7.
14. Dasgupta, A.; Chowdhury, N.; De, R. K. Comput. Methods Programs Biomed. 2020, 192, 105436. DOI: https://doi.org/10.1016/j.cmpb.2020.105436.
15. Marchev, A. S.; Yordanova, Z. P.; Georgiev, M. I. Crit. Rev. Biotechnol. 2020, 40, 443–458. DOI: https://doi.org/10.1080/07388551.2020.1731414.
16. Yamada, Y.; Sato, F. Biomolecules. 2021, 11, 1719. DOI: https://doi.org/10.3390/biom11111719.
17. Xool-Tamayo, J.; Serrano-Gamboa, G.; Monforte-González, M.; Mirón-López, G.; Vázquez-Flota, F. Biotechnol. Lett. 2017, 39, 323–330. DOI: https://doi.org/10.1007/s10529-016-2250-9.
18. http://www.bioinformatics.babraham.ac.uk/projects/fastqc, accessed in October 2023.
19. http://hannonlab.cshl.edu/fastx_toolkit, accessed in January 2024.
20. Schulz, M. H.; Zerbino, D. R.; Vingron, M.; Birney, E. Bioinformatics. 2012, 28, 1086–1092. DOI: https://doi.org/10.1093/bioinformatics/bts094.
21. Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E. V.; Zdobnov, E.M. Bioinformatics, 2015, 31, 3210-3212. DOI: https://doi.org/10.1093/bioinformatics/btv351.
22. Grabherr, M. G.; Haas, B. J.; Yassour, M.; Levin, J. Z.; Thompson, D. A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; Chen, Z.; Mauceli, E.; Hacohen, N.; Gnirke, A.; Rhind, N.; di Palma, F.; Birren, B. W.; Nusbaum, C.; Lindblad-Toh, K.; Friedman, N.; Regev, A. Nat. Biotechnol. 2011, 29, 644–652. DOI: https://doi.org/10.1038/nbt.1883.
23. Danecek, P.; Bonfield, J. K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M. O.; Whitwham, A.; Keane, T.; McCarthy, S. A.; Davies, R. M.; Li, H. GigaScience. 2021, 10, giab008. DOI: https://doi.org/10.1093/gigascience/giab008.
24. Quinlan, A. R.; Hall, I. M. Bioinformatics. 2010, 26, 841–842. DOI: https://doi.org/10.1093/bioinformatics/btq033
25. Eddy, S. R. PLoS Computational Biology. 2011, 7, e1002195. DOI: https://doi.org/10.1371/journal.pcbi.1002195.
26. Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G. A.; Sonnhammer, E. L. L.; Tosatto, S. C. E.; Paladin, L.; Raj, S.; Richardson, L. J.; Finn, R. D.; Bateman, A. Nucleic Acids Res. 2021, 49, D412–D419. DOI: https://doi.org/10.1093/nar/gkaa913.
27. Dastmalchi, M.; Park, M. R.; Morris, J. S.; Facchini, P. Phytochem. Rev. 2018, 17, 249–277. DOI: https://doi.org/10.1007/s11101-017-9519-z.
28. Gracz‐bernaciak, J.; Mazur, O.; Nawrot, R. Int. J. Mol. Sci. 2021, 22, 12427. DOI: https://doi.org/10.3390/ijms222212427.
29. Facchini, P. J.; Morris, J. S. Front. Plant Sci. 2019, 10, 1058. DOI: https://doi.org/10.3389/fpls.2019.01058.
30. Zhong, F.; Huang, L.; Qi, L.; Ma, Y.; Yan, Z. Plant Mol. Biol. 2020, 102, 477–499. DOI: https://doi.org/10.1007/s11103-019-00959-y.
31. Buchfink, B.; Reuter, K.; Drost, H.-G. Nat. Methods. 2021, 18, 366–368. DOI: https://doi.org/10.1038/s41592-021-01101-x.
32. Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. Bioinformatics. 2020, 36, 2251–2252. DOI: https://doi.org/10.1093/bioinformatics/btz859.
33. Kanehisa, M.; Sato, Y. Protein Sci. 2020, 29, 28–35. DOI: https://doi.org/10.1002/pro.3711.
34. Törönen, P.; Medlar, A.; Holm, L. Nucleic Acids Res. 2018, 46, W84–W88. DOI: https://doi.org/10.1093/nar/gky350.
35. Jones D.T.; Taylor W.R. FEBS Lett 1994, 339, 269–275. DOI:10.1016/0014-5793(94)80429-X.
36. Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. PLoS ONE. 2011, 6, e21800. DOI: https://doi.org/10.1371/journal.pone.0021800.
37. Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. Mol. Biol. Evol. 2018, 35, 1547–1549. DOI: https://doi.org/10.1093/molbev/msy096.
38. Kelley, L. A.; Mezulis, S.; Yates, C. M.; Wass, M. N.; Sternberg, M. J. E. Nat. Protoc. 2015, 10, 845–858. DOI: https://doi.org/10.1038/nprot.2015.053.
39. Vogel, M.; Lawson, M.; Sippl, W.; Conrad, U.; Roos, W. J. Biol. Chem. 2010, 285, 18397–18406. DOI: https://doi.org/10.1074/jbc.M109.088989.
40. Deng, X.; Zhao, L.; Fang, T.; Xiong, Y.; Ogutu, C.; Yang, D.; Vimolmangkang, S.; Liu, Y.; Han, Y. Hortic. Res. 2018, 5, 29. DOI: https://doi.org/10.1038/s41438-018-0035-0.
41. Hagel, J. M.; Morris, J. S.; Lee, E.-J.; Desgagné-Penix, I.; Bross, C. D.; Chang, L.; Chen, X.; Farrow, S. C.; Zhang, Y.; Soh, J.; Sensen, C. W.; Facchini, P. J. BMC Plant Biol. 2015, 15, 227. DOI: https://doi.org/10.1186/s12870-015-0596-0.
42. Pei, L.; Wang, B.; Ye, J.; Hu, X.; Fu, L.; Li, K.; Ni, Z.; Wang, Z.; Wei, Y.; Shi, L.; Zhang, Y.; Bai, X.; Jiang, M.; Wang, S.; Ma, C.; Li, S.; Liu, K.; Li, W.; Cong, B. Hortic. Res. 2021, 8, 5. DOI: https://doi.org/10.1038/s41438-020-00435-5.
43. Morris, J. S.; Caldo, K. M. P.; Liang, S.; Facchini, P. J. ChemBioChem. 2021, 22, 264–287. DOI: https://doi.org/10.1002/cbic.202000354.
44. Hagel, J. M.; Beaudoin, G. A. W.; Fossati, E.; Ekins, A.; Martin, V. J. J.; Facchini, P. J. J. Biol. Chem. 2012, 287, 42972–42983. DOI: https://doi.org/10.1074/jbc.M112.420414.
45. Díaz Chávez, M. L.; Rolf, M.; Gesell, A.; Kutchan, T. M. Arch. Biochem. Biophys. 2011, 507, 186–193. DOI: https://doi.org/10.1016/j.abb.2010.11.016
46. Leong, B. J.; Last, R. L. Curr. Opin. Struct. Biol. 2017, 47, 105–112. DOI: https://doi.org/10.1016/j.sbi.2017.07.005
47. Waki, T.; Takahashi, S.; Nakayama, T. BioEssays. 2021, 43, 2000164. DOI: https://doi.org/10.1002/bies.202000164
48. Loza-Muller, L.; Shitan, N.; Yamada, Y.; Vázquez-Flota, F. Planta. 2021, 254, 122. DOI: https://doi.org/10.1007/s00425-021-03780-4
49. Kato, N.; Dubouzet, E.; Kokabu, Y.; Yoshida, S.; Taniguchi, Y.; Dubouzet, J. G.; Yazaki, K.; Sato, F. Plant Cell Physiol. 2007, 48, 8–18. DOI: https://doi.org/10.1093/pcp/pcl041.
50. Yamada, Y.; Kokabu, Y.; Chaki, K.; Yoshimoto, T.; Ohgaki, M.; Yoshida, S.; Kato, N.; Koyama, T.; Sato, F. Plant Cell Physiol. 2011, 52, 1131–1141. DOI: https://doi.org/10.1093/pcp/pcr062.

Downloads
Published
Issue
Section
License
Copyright (c) 2025 José Germán Serrano-Gamboa, Jorge Froylán Xool-Tamayo, Lloyd Loza-Muller, Yahaíra de Jesús Tamayo-Ordóñez, Felipe A. Vázquez-Flota

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.