Theoretical Investigation of the Cooperation of Iminoguanidine with the Enzymes-Binding Domain of Covid-19 and Bacterial Lysozyme Inhibitors and their Pharmacokinetic Properties
Iminoguanidine Derivatives as Multi-target Lead Compound Against Covid-19 and Pseudomonas aeruginosa
DOI:
https://doi.org/10.29356/jmcs.v66i4.1726Keywords:
Pseudomonas aeruginosa, SARS-CoV-2, QSAR, homology modelling, Ramachandran plot, docking simulations, molecular dynamic simulations, ADMEAbstract
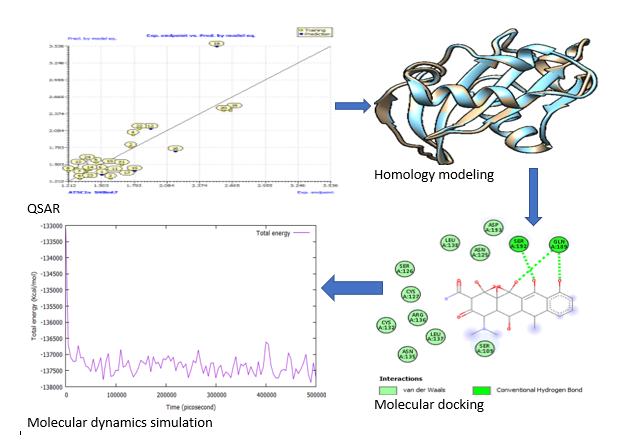
Abstract. The investigation for innovative treatments for Pseudomonas aeruginosa and SARS-CoV-2 is a burgeoning field. Even though scientists and pharmaceutical companies have made significant contributions to the research of multi-drug resistance infections from a variety of perspectives, these diseases remain incurable. As a result, developing novel and more effective drugs for proteins associated with Pseudomonas aeruginosa and SARS-CoV-2 has become a top priority in recent years. In this regard, the protein is known as a putative inhibitor of vertebrate lysozyme [Pseudomonas aeruginosa] and chain A, spike protein S1 [SARS-CoV-2], and it is one of the key targets for the development of new drug candidates that could be used as inhibitors in both Pseudomonas aeruginosa and SARS-CoV-2 chemotherapies. The structural characteristics and binding mechanism of certain inhibitors of Pseudomonas aeruginosa and SARS-CoV-2 receptor were investigated using Quantitative-Structure Activity Relationship (QSAR), homology modeling, molecular docking, and molecular dynamics simulation in this study. To create a 3D model of the receptors, a homology modeling approach was used. The X-ray crystal structures of chain A inhibitor of vertebrate lysozyme (PDB: 4PS6) and chain A spike protein S1 (PDB: 7MZF), respectively, were employed as templates for this technique. The final projected structure is obtained and examined by the programs ERRAT, VERIFY3D, and PROCHECK, confirming that the final model is credible. The anticipated structure was fine-tuned before being employed in docking simulations. The results of the structure-based virtual screening show that two potent new lead molecules, compounds 7 and 15, have the most noteworthy affinity to the predicted 3D receptors. The docked compound 15 was subjected to a 1 ns molecular dynamics (MD) simulation. Compound 15 produced more hydrophobic and van der Waal's contacts, according to MD simulations, and binds to SARS-CoV-2 spike protein S1 more closely. The Lipinski rule of five assessment revealed that these lead compounds had significant pharmacological characteristics. As a result, the current research will aid in the development and synthesis of another class of chain A inhibitors of vertebrate lysozyme and chain A spike protein S1 inhibitors that restore drug compound susceptibilities.
Resumen. La investigación de tratamientos innovadores para Pseudomonas aeruginosa y SARS-CoV-2 es un campo floreciente. A pesar de que los científicos y las compañías farmacéuticas han hecho contribuciones significativas a la investigación de infecciones por resistencia a múltiples medicamentos desde una variedad de perspectivas, estas enfermedades siguen siendo incurables. Como resultado, el desarrollo de fármacos novedosos y más eficaces para las proteínas asociadas con Pseudomonas aeruginosa y SARS-CoV-2 se ha convertido en una prioridad en los últimos años. En este sentido, la proteína es conocida como un supuesto inhibidor de la lisozima de vertebrados [Pseudomonas aeruginosa] y de la cadena A, proteína espiga S1 [SARS-CoV-2], y es uno de los objetivos clave para el desarrollo de nuevos fármacos candidatos que podrían utilizarse como inhibidores tanto en pseudomonas aeruginosa como en quimioterapias de SARS-CoV-2. Las características estructurales y el mecanismo de unión de ciertos inhibidores de Pseudomonas aeruginosa y el receptor SARS-CoV-2 se investigaron utilizando la Relaciones Cuantitativas de Estructura-Actividad - (QSAR, por sus siglas en inglés), el modelado por homología, el acoplamiento molecular y la simulación de dinámica molecular. Para crear un modelo 3D de los receptores, se utilizó un enfoque de modelado por homología. Las estructuras cristalinas de rayos X del inhibidor de la cadena A de la lisozima de vertebrados (PDB: 4PS6) y la proteína de espiga de cadena A S1 (PDB: 7MZF), respectivamente, se emplearon como plantillas para esta técnica. La estructura final proyectada se obtuvo y examinó con los programas ERRAT, VERIFY3D y PROCHECK, confirmando que el modelo final es creíble. La estructura anticipada se afinó antes de ser empleada en simulaciones de acoplamiento. Los resultados del cribado virtual basado en la estructura generaron dos nuevas y potentes moléculas líderes, los compuestos 7 y 15, que tienen la afinidad más notable con los receptores 3D predichos. El compuesto acoplado 15 fue sometido a una simulación de dinámica molecular (DM) de 1 ns. El compuesto 15 produjo más contactos hidrófobos y de van der Waals, según las simulaciones de MD, y se une más de cerca a la proteína espiga S1 del SARS-CoV-2. La evaluación de la regla de cinco de Lipinski reveló que estos compuestos líderes tenían características farmacológicas significativas. Como resultado, la investigación actual ayudará en el desarrollo y la síntesis de otra clase de inhibidores de la cadena A de la lisozima de vertebrados y los inhibidores de la proteína de espiga de cadena A S1 que restauran las susceptibilidades de los compuestos farmacológicos.
Downloads
References
Edache, E. I.; Uzairu, A.; Mamza, P. A. P.; Shallengwa, G. A. J. Drug. Design. Discov. Res. 2020, 1, 36-52.
Yuan, M.; Guan, H.; Sha. D.; Cao, W.; Song, X.; Che, J.; Kan, B.; Li, J. Antibiotics. 2021, 10, 1234. DOI: https://DOI.org/10.3390/antibiotics10101234.
Azam, M. W.; Khan, A. U. Drug Discov. Today. 2019, 24, 350–359. DOI: https://doi.org/10.1016/j.drudis.2018.07.003.
Sahu, M. K.; Kaushik, K.; Das, A.; Jha, H. Bioresour. Bioprocess. 2020, 7, 14. DOI: https://doi.org/10.1186/s40643-020-00303-z.
Ahlgren, H. G.; Benedetti, A.; Landry, J. S.; Bernier, J.; Matouk, E.; Radzioch, D.; Lands, L. C.; Rousseau, S.; Nguyen, D. BMC Pulm. Med. 2015, 15, 67. https://doi.org/10.1186/s12890-015-0062-7.
Gifford, A. H.; Miller, S. D.; Jackson, B. P.; Hampton, T. H.; O’Toole, G. A.; Stanton, B. A.; Parker, H. W. Pediatr. Pulmonol. 2011, 46, 160-165. DOI: https://doi.org/10.1002/ppul.21335.
Callewaert, L.; Aertsen, A.; Deckers, D.; Vanoirbeek, K. G. A.; Vanderkelen L, et al. PLoS Pathog. 2008, 4, e1000019. DOI: https://doi.org/10.1371/journal.ppat.1000019.
Callewaert, L.; Van Herreweghe, J. M.; Vanderkelen, L.; Leysen, S.; Voet, A.; Michiels, C. W. Trends Microbiol. 2012, 20, 501-510. DOI: https://doi.org/10.1016/j.tim.2012.06.005.
Yum, S.; Kim, M. J.; Xu, Y.; Jin. X. L.; Yoo. H. Y.; Park, J. W.; Gong, J. H.; Choe, K. M.; Lee, B. L.; Ha, N. C. Biochem Biophys Res Commun. 2009, 378, 244-248. DOI: https://doi.org/10.1016/j.bbrc.2008. DOI: https://doi.org/10.1016/j.bbrc.2008.11.039
Leysen, S.; Van Herreweghe J. M.; Callewaert, L.; Heirbaut, M.; Buntinx, P.; Michiels, C. W.; Strelkov, S. V. J. Mol. Bio. 2011, 405, 1233–1245. DOI: 10.1016/j.jmb.2010.12.007. DOI: https://doi.org/10.1016/j.jmb.2010.12.007
Li, Y.; Li, H.; Fan, R.; Wen, B.; Zhang, J.; Cao, X.; Wang, C.; Song, Z.; Li, S.; Li, X.; Lv, X. Intervirology. 2016, 59, 163-169. DOI: https://doi.org/10.1016/j.jmb.2010.12.007. DOI: https://doi.org/10.1159/000453066
Noman, A.; Aqeel, M.; Khalid, N.; Hashem, M.; Alamari, S.; Zafar, S.; Qasim, M.; Irshad, M. K.; Qari, S. H. Microb. Pathog. 2021, 150, 104719. DOI: https://doi.org/10.1016/j.micpath.2020.104719.
Chan. J. F. W.; Yuan, S.; Kok, K. H.; To, K. K. W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C. C. Y.; Poon, R. W. S.; Tsoi, H. W. Lancet. 2020, 395, 514-523. DOI: https://doi.org/10.1016/S0140-6736(20)30154-9.
Zou, L.; Ruan, F.; Huang, M.; Liang, L.; Huang, H.; Hong, Z.; Yu, J.; Kang, M.; Song, Y.; Xia, J.; Guo, Q. New Eng. J. Med. 2020, 382, 1177-1179. DOI: https://doi.org/10.1056/NEJMc2001737.
Rhoades, N. S.; Pinski, A. N.; Monsibais, A. N.; Jankeel, A.; Doratt, B. M.; Cinco, I. R. Cell Rep. 2021, 36, 109637. DOI: https://doi.org/10.1016/j.celrep.2021.109637.
Simmons, G.; Reeves, J. D.; Rennekamp, A. J.; Amberg, S. M.; Piefer, A. J.; Bates, P. Proc. Natl. Acad. Sci. 2004, 101, 4240–4245. DOI: https://doi.org/10.1073/pnas.0306446101.
Guo, Y. R.; Cao, Q. D.; Hong, Z. S.; Tan, Y. Y.; Chen, S. D.; Jin, H. J.; Tan, K. S.; Wang, D. Y.; Yan, Y. Military Med. Res. 2020, 7, 1-10. DOI: https://doi.org/10.1186/s40779-020-00240-0.
Condé, K.; Atakla, H. G.; Garba, M. S.; Garba, I. Afr. Med. J. 2020, 35, 96. DOI: https://doi.org/10.11604/pamj.supp.2020.35.2.24616. DOI: https://doi.org/10.11604/pamj.supp.2020.35.2.24549
Kim, S. Y.; Jin, W.; Sood, A.; Montgomery, D. W.; Grant, O. C.; Fuster, M. M.; Fu, L.; Dordick, J. S.; Woods, R. J.; Zhang, F.; Linhardt, R. J. Antiviral Res. 2020, 181, 104873. DOI: https://DOI.org/10.1016/j.antiviral.2020.104873.
Muratov, E. N.; Amaro, R.; Andrade, C. H.; Brown, N.; Ekins, S.; Fourches, D.; Isayev, O.; Kozakov, D.; et al. Chem. Soc. Rev. 2021, 50, 9121-9151. DOI: https://doi.org/10.1039/D0CS01065K.
Tortorici, M. A.; Walls, A. C.; Lang, Y.; Wang, C.; Li, Z.; Koerhuis, D.; Boons, G. J.; Bosch, B. J.; Rey, F. A.; de Groot, R. J.; Veesler, D. Nat. Struct. Mol. Biol. 2019, 26, 481–489. https://DOI.org/10.1038/s41594-019-0233-y.
Vashi, Y.; Jagrit, V.; Kumar, S. Infec., Gen. Evol. 2020, 84, 104382. DOI: https://DOI.org/10.1016/j.meegid.2020.104382.
Kanaan, M. H. G. Vet. World. 2018, 11, 1445-1453. DOI: https://doi.org/10.14202/vetworld.2018.1445-1453.
Heinzl, G. A.; Huang, W.; Yu, W.; Giardina, B. J.; Zhou, Y.; MacKerell, Jr., A. D.; Wilks, A.; and Xue, F. J Med Chem. 2016, 59, 6929-6942. DOI: https://doi.org/10.1021/acs.jmedchem.6b00757.
Edache, E. I.; Uzairu, A.; Mamza, P. A.; Shallangwa, G. A. J. Genet. Eng. Biotechnol. 2022, 20, 88. DOI: https://doi.org/10.1186/s43141-022-00362-z.
Edache, E. I.; Uzairu, A.; Mamza, P. A.; Shallangwa, G. A. J. Virol. Viral Dis. 2022, 2, 1-28. DOI: https://doi.org/10.54289/JVVD2200106.
Custelcean, R. Chem. Commun. 2020, 56, 10272-10280. DOI: https://doi.org/10.1039/D0CC04332J.
Zhang Q.; Jiang, Y.; Li, Y.; Song, X.; Luo, X.; Ke, Z.; and Zou, Y. iScience. 2021, 24, 102263. DOI: https://doi.org/10.1016/j.isci.2021.102263.
Maia, E. H. B.; Assis, L. C.; Oliveira, T. A.; Silva, A. M.; Taranto, A. G. Front. Chem. 2020, 8, 343. DOI: https://doi.org/10.3389/fchem.2020.00343.
Liu, Y.; Zhang, L.; Guo, M.; Wu, H.; Xie, J.; Wei, D. Bioresour. Bioprocess. 2014, 1,1-9. DOI: https://doi.org/10.1186/s40643-014-0017-5.
Shirvani, P.; Fassihi, A. Mol. Simul. 2020, 46, 16, 1265-1280. DOI: https://doi.org/10.1080/08927022.2020.1810853.
Becke, A. D. J. Chem. Phys. 1993, 98, 5648-5652. DOI: https://doi.org/10.1063/1.464913.
Lee, C.; Yang, W.; Parr, R. G. Phys. Rev. B. 1988, 37, 785-789. DOI: https://doi.org/10.1103/physrevb.37.785.
Petersson, G. A.; Bennett, A.; Tensfeldt, T. G.; Al-Laham, M. A.; Shirley, W. A.; Mantzaris, J. J. Chem. Phys. 1988, 89, 2193-2218. DOI: https://doi.org/10.1063/1.455064.
Frisch, M. J.; Trucks, G.W.; Schlegel, H. B.; Scuseria, G. E.; Robb, M. A.; Cheeseman, J. R.; Scalmani, G.; Barone, V.; Petersson, G. A.; Nakatsuji, H.; et al. Revision A.02. 2016, Gaussian Inc, Wallingford CT
Yap, C. W. J. Comput. Chem. 2011, 32, 1466-1474. DOI: https://doi.org/10.1002/jcc.21707.
Gramatica, P.; Chirico, N.; Papa, E.; Cassani, S.; Kovarich, S. J. Comput. Chem. 2013. DOI: https://doi.org/10.1002/jcc.23361.
Edache, E. I.; Uzairu, A.; Mamza, P. A.; Shallangwa, G. A. Sci. Afr. 2021, 15, e01088. DOI: https://doi.org/10.1016/j.sciaf.2021.e01088.
Roy, K.; Mitra, I. Comb. Chem. High Throughput Screen. 2011, 14, 450–474. DOI: https://doi.org/10.2174/138620711795767893.
Roy, K.; Chakraborty, P.; Mitra, I.; Ojha, P. K.; Kar, S.; Das, R. N. J. Comput. Chem. 2013, 34, 1071-1082. DOI: https://doi.org/10.1002/jcc.23231.
Edache, E. I.; Uzairu, A.; Abechi, S. E. J. Comput. Methods Mol. Des. 2015, 5, 135-149
Sneath, P. H.; Sokal, R. R. Numerical Taxonomy San Francisco. A statistical method for evaluating systematic relationships, 1973, 38, 1409-1438.
Zuckerkandl, E.; Pauling, L. in: Evolutionary divergence and convergence in proteins. In Evolving genes and proteins, Academic Press., 1965, 97-166. DOI: https://doi.org/10.1016/B978-1-4832-2734-4.50017-6
Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. J. Mol. Biol. Evol. 2018, 35, 1547-1549. DOI: https://doi.org/10.1093/molbev/msy096.
Li, Y.; Qiu, Y.; Zhang, X.; Zhu, M.; Tan, W. Bioresour. Bioprocess. 2019, 6, 15. DOI: https://DOI.org/10.1186/s40643-019-0250-z.
Sali, A.; Blundell, T. L. J. Mol. Biol. 1993, 234, 779-815. DOI: https://doi.org/10.1006/jmbi.1993.1626.
Feng, J.; Xu, S.; Feng, R.; Kovalevsky, A.; Zhang, X.; Liu, D.; Wan, Q. Bioresour Bioprocess. 2021, 8, 102. DOI: https://doi.org/10.1186/s40643-021-00449-4.
Hall, T. A. Nucl. Acids. Symp. Ser. 1999, 41, 95-98. DOI: https://doi.org/10.1099/00222615-48-1-95
Guex, N.; Peitsch, M. C. Electrophoresis 1997, 18, 2714-2723. DOI: https://doi.org/10.1002/elps.1150181505.
Thomsen, R.; Christensen, M. H. Molegro Virtual Docker 4.0 User Manual; Molegro ApS: Aarhus, Denmark, 2009, 125-133.
Dallakyan, S.; Olson, A. J. Methods Mol. Biol. 2015, 1263, 243–250. DOI: https://doi.org/10.1007/978-1-4939-2269-7_19.
Trott, O.; Olson, A. J. J. Comput. Chem. 2010, 31, 455–461. DOI: https://doi.org/10.1002/jcc.21334.
Jo, S.; Kim, T.; Iyer, V. G.; Im, W. J. Comput. Chem. 2008, 29, 1859–1865. DOI: https://doi.org/10.1002/jcc.20945.
Lee, J.; Cheng, X.; Swails, J. M.; Yeom, M. S.; Eastman, P. K.; Lemkul, J. A.; Wei, S.; Buckner, J.; Jeong, J. C.; Qi, Y, Jo, S. J. Chem. Theory Comput. 2016, 12, 405-413. DOI: https://doi.org/10.1021/acs.jctc.5b00935.
Phillips, J. C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R. D.; Kale, L.; Schulten, K. J. Comput. Chem. 2005, 26, 1781-1802. DOI: https://doi.org/10.1002/jcc.20289.
Humphrey, W.; Dalke, A.; Schulten, K. J. Mol. Graphics. 1996, 14, 33-38.51. DOI: https://doi.org/10.1016/0263-7855(96)00018-5.
El-Mernissi, R.; El Khatabi K.; Khaldan, A.; ElMchichi, L.; Shahinozzaman, M.; Mohammed Aziz Ajana, M. A.; Lakhlifi, T.; Bouachrine, M. J. Mex. Chem. Soc. 2022, 66, 79-94. DOI: http://dx.doi.org/10.29356/jmcs.v66i1.1578. DOI: https://doi.org/10.29356/jmcs.v66i1.1578
Bolboac, S. D.; Lorentz, J. Biomath. 2013, 2, 1-11. DOI: http://dx.doi.org/10.11145/j.biomath.2013.09.089. DOI: https://doi.org/10.11145/j.biomath.2013.09.089
Edache, E. I.; Uzairu, A.; Mamza, P. A.; Shallangwa, G. A. Biomed. J. Sci. Tech. Res. 2021, 34. 26472- 26489. DOI: https://doi.org/10.26717/BJSTR.2021.34.005509.
Tropsha, A.; Gramatica, P.; Gombar, V. K. QSAR Comb. Sc. 2003, 22, 69-77. DOI: https://doi.org/10.1002/qsar.200390007.
Ruiz, I. L.; Gomez-Nieto, M. A. Molecules. 2018, 23, 2756. DOI: https://doi.org/10.3390/molecules23112756.
Gramatica, P.; Cassani, S.; Chirico, N. J. Comput. Chem. 2014, 35, 1036–1044. DOI: https://doi.org/10.1002/jcc.23576.
Hall, B. G. Mol. Biol. Evol. 2013, 30, 1229-1235. https://doi.org/10.1093/molbev/mst012.
Edgar, R. C. BMC Bioinf. 2004, 5, 113. DOI: https://doi.org/10.1186/1471-2105-5-113.
Elengoe, A.; Naser, M. A.; Hamdan, S. Biologia. 2015, 70, 1655-1671. DOI: https://doi.org/10.1515/biolog-2015-0194.
Mugabo, B.; Iradukunda, R.; Gatanganwa, J. M.; Ufitinema, J. C.; Mutangana, D. Rwanda J. Engr. Sci. Tech. Environ. 2021, 4. DOI: https://dx.DOI.org/10.4314/rjeste.v4i1.3. DOI: https://doi.org/10.4314/rjeste.v4i1.3
Tambunan, U. S. F.; Bramantya, N.; Parikesit, A. A. BMC Bioinf. 2011, 12, S23. DOI: http://www.biomedcentral.com/1471-2105/12/S13/S23. DOI: https://doi.org/10.1186/1471-2105-12-S13-S23
Chen, J.; Luo, X. J.; Chen, Q.; Pan, J.; Zhou, J.; Xu, J. H. Bioresour. Bioprocess. 2015, 2, 39. DOI: https://doi.org/10.1186/s40643-015-0067-3.
Thomsen, R.; Christensen, M. H. J. Med. Chem. 2006, 49, 3315-3321. DOI: https://doi.org/10.1021/jm051197e.
Lipinski, C. A.; Lombardo, F.; Dominy, B. W.; Feeney, P. J. Adv. Drug. Deliv. Rev. 2001, 46, 3–26. DOI: https://doi.org/10.1016/s0169-409x(00)00129-0.
Edache, E. I.; Uzairu, A.; Shallangwa, G. A.; Mamza, P. A. Future J. Pharm. Sci. 2021, 7, 1-22. DOI: https://doi.org/10.1186/s43094-021-00367-4.

Downloads
Additional Files
Published
Issue
Section
License
Copyright (c) 2022 Emmanuel Israel Edache, Adamu Uzairu, Paul Andrew Mamza, Gideon Adamu Shallangwa

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.