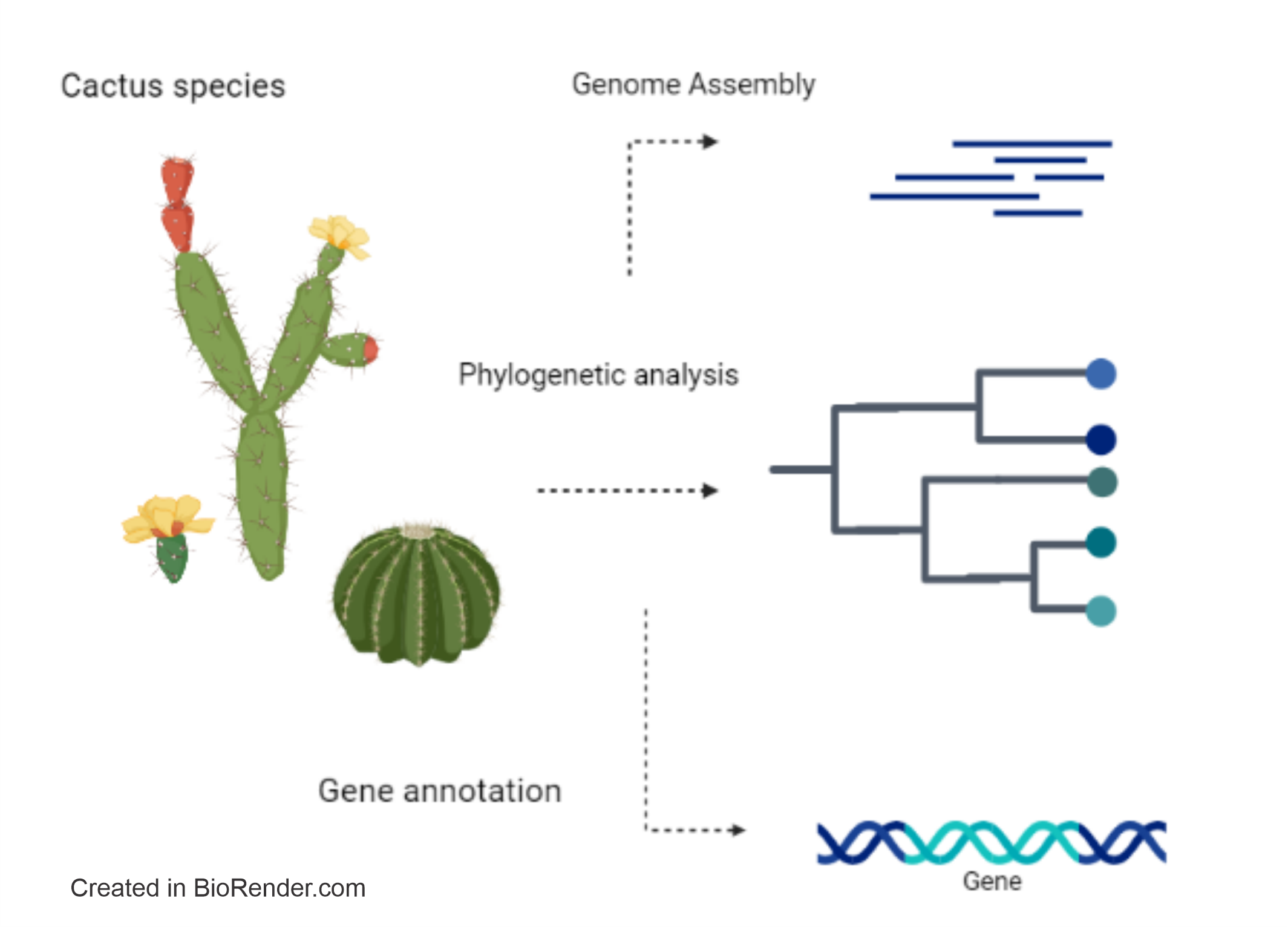
Assembly of the Cereus fernambucensis Genome, Gene Annotation, and Tertiary Structure of Secondary Metabolism Enzymes in Carnegiea gigantea, Lophocereus schottii, Pachycereus pringlei, Pereskia humboldtii, Selenicereus undatus, and Stenocereus thurberi
DOI:
https://doi.org/10.29356/jmcs.v67i3.1969Keywords:
Cactaceae, genome assembly, gene annotation, secondary metabolism, bioactive compoundsAbstract
Abstract. Recently, there is growing interest in obtaining bioactive compounds from species in the family Cactaceae, which has been little analyzed at the genomic and transcriptomic level. We here report the assembly of the genome of Cereus fernambucensis and we analyzed six cactus genomes (Carnegiea gigantea, Lophocereus schottii, Pachycereus pringlei, Pereskia humboldtii, Selenicereus undatus and Stenocereus thurberi), the annotation of putative genes, and the modeling of the three-dimensional structures of their predicted proteins involved in flavonoid metabolism. We identified genes encoding proteins related to plant pathogenesis (PR-10), coding secuences (CDS) of aldehyde reductase and flavonoid reductase, CDS of enzymes involved in the biosynthesis of phenolic compounds, and ABC transporters. The grouping of the enzymes aspartic proteinase-like protein, flavanone 3-hydroxylase (F3H), hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase (HCT), and protein serine/threonine- phosphatase was shown to be highly conserved in the genomes of the analyzed cacti. We found divergence of the plant PDR ABC-type transporter family protein (PEN3) in Cereus fernambucensis and the absence in this species of sterol methyltransferase (SMT1). Our three-dimensional modeling of the tertiary structure of F3H from a consensus sequence of cactus species had 88 % identity with that reported in Arabidopsis thaliana. We observed the conservation in several plant species of the 2-oxoglutarate and iron-dependent domain of F3H. This is the first report of an exploration of putative genes encoding enzymes involved in secondary metabolism in cacti species providing information that could be used to improve the production of bioactive compounds in them.
Resumen. Recientemente, ha incrementado el interés en obtener compuestos bioactivos de especies de la familia Cactaceae, que ha sido poco analizada a nivel genómico y transcriptómico. Nosotros reportamos el ensamblaje del genoma Cereus fernambucensis y analizamos además seis genomas de cactus (Carnegiea gigantea, Lophocereus schottii, Pachycereus pringlei, Pereskia humboldtii, Selenicereus undatus y Stenocereus thurberi), la anotación de genes putativos y el modelado de las estructuras tridimensionales de sus proteínas involucradas en el metabolismo de los flavonoides. Se identificaron genes que codifican proteínas relacionadas con la patogénesis vegetal (PR-10), secuencias codificantes (CDS) de aldehído reductasas y flavonoide reductasas, CDS de enzimas implicadas en la biosíntesis de compuestos fenólicos y transportadores ABC. La agrupación de las enzimas similar a la proteinasa aspártica, flavanona 3-hidroxilasa (F3H), hidroxicinamoil-CoA shikimato/quinato hidroxicinamoiltransferasa (HCT) y proteína serina/treonina-fosfatasa demostró estar altamente conservada en los genomas de los cactus analizados. Se encontró divergencia de la proteína (PEN3) de la familia transportadora de tipo ABC PDR en Cereus fernambucensis y la ausencia en esta especie de esterol metiltransferasa (SMT1). Nuestro modelado tridimensional de la estructura terciaria de F3H a partir de una secuencia consenso de especies de cactus tuvo una identidad del 88 % con la reportada en Arabidopsis thaliana. Observamos la conservación en varias especies vegetales del dominio 2-oxoglutarato y dependiente del hierro de F3H. Este es el primer informe de una exploración de genes putativos que codifican enzimas involucradas en el metabolismo secundario en especies de cactus que aporta información que podría usarse para mejorar la producción de compuestos bioactivos.
Downloads
References
Guerrero, P.C.; Majure, L.C.; Cornejo-Romero, A.; Hernández-Hernández, T.J. Hered. 2019, 110, 4-21. DOI:10.1093/jhered/esy064. DOI: https://doi.org/10.1093/jhered/esy064
Casas A.; Barbera G., Cacti: Biology and Uses. Ed., Park S. Nobel, 2002, 142-162. DOI: 10.1525/california/9780520231573.003.0009. DOI: https://doi.org/10.1525/california/9780520231573.003.0009
Figueroa, R.; Tamayo, J.; González, S.; Moreno, G.; Vargas, L. Revista Iberoamericana de Tecnología Postcosecha. 2011, 12, 44-50. DOI: redalyc.org/articulo.oa?id=81318808007
Bargougui, A.; Tag, H. M.; Bouaziz, M.; Triki, S. Biomed. Pharmacol. J. 2019, 12, 1353-1368. DOI: https://dx.doi.org/10.13005/bpj/1764 DOI: https://doi.org/10.13005/bpj/1764
Cornejo-Campos, J.; Gómez-Aguirre, Y.A.; Velázquez-Martínez, J.R.; Ramos-Herrera, O.J.; Chávez-Murillo, C.E.; Cruz-Sosa, F.; Areche, C.; Cabañas-García, E. Molecules. 2022, 27, 3707. DOI: 10.3390/molecules27123707. DOI: https://doi.org/10.3390/molecules27123707
Fernández-López, J.A.; Almela, L.; Obón, J.M.; Castellar, R. Plant Foods Hum. Nutr. 2010, 65, 253-259. DOI: 10.1007/s11130-010-0189-x. DOI: https://doi.org/10.1007/s11130-010-0189-x
Torres-Silva, G.; Correia, L. N. F.; Batista, D. S.; Koehler, A.D.; Resende, S.V.; Romanel, E.; Cassol, D.; Rocha-Almeida, A.M.; Strickler, S.R.; Dvorak-Specht, C.; Otoni, W.C. Front. Plant Sci. 2021, 1658. DOI: https://doi.org/10.3389/fpls.2021.697556.
Enríquez-González, C.; Garcidueñas-Piña, C.; Castellanos-Hernández, O. A.; Enríquez-Aranda, S.; Loera-Muro, A.; Ocampo, G.; Pérez-Molphe Balch, E.; Morales-Domínguez, J.F. Plants. 2022, 11, 399. DOI: 10.3390/plants11030399. DOI: https://doi.org/10.3390/plants11030399
Ibarra-Laclette, E.; Zamudio-Hernández, F.; Pérez-Torres, C.A.; Albert, V.A.; Ramírez-Chávez, E.; Molina-Torres, J.; Férnandez-Cortes, A.; Calderón-Vázquez, C.; Olivares-Romero, J.L.; Herrera-Estrella, A.; Herrera-Estrella, L. BMC genomics. 2015, 16, 1-14. DOI 10.1186/s12864-015-1821-9 DOI: https://doi.org/10.1186/s12864-015-1821-9
Sykłowska-Baranek, K.; Kamińska, M.; Pączkowski, C.; Pietrosiuk, A.; Szakiel, A. Plants. 2022, 11, 1120. DOI: 10.3390/plants11091120. DOI: https://doi.org/10.3390/plants11091120
Yan, D.; Tajima, H.; Cline, L.C.; Fong, R.Y.; Ottaviani, J.I.; Shapiro, H.Y.; Blumwald, E. Plant biotechnol. J. 2022, 20, 2135-2148. DOI: https://doi.org/10.1111/pbi.13894.
Zheng, J.; Meinhardt, L.W.; Goenaga, R.; Zhang, D.; Yin, Y. Hortic. Res. 2021, 8. DOI: https://doi.org/10.1038/s41438-021-00501-6.
Franco, F.F.; Amaral, D.T.; Bonatelli, I.A.; Romeiro-Brito, M.; Telhe, M.C.; Moraes, E.M. Genes. 2022, 13, 452. DOI: 10.3390/genes13030452. DOI: https://doi.org/10.3390/genes13030452
Oliveira, de A.; Gabrielly, M.; Morghanna Barbosa do Nascimento, G.; Ribeiro Orasmo, G., in: Wild Plants. CRC Press, 2020, 326-337.
Orozco-Barocio, A.; Robles-Rodríguez, B.S.; del Rayo Camacho-Corona, M.; Méndez-López, L.F.; Godínez-Rubí, M.; Peregrina-Sandoval, J.; Rivera, J.; Rojas Mayorquín, A.E.; Ortuno-Sahagun, D. Front. Pharmacol. 2022, 13. DOI: https://doi.org/10.3389/fphar.2022.820381.
Lin, X.E.; Gao, H.; Ding, Z.; Zhan, R.; Zhou, Z.; Ming, J. BioMed Res. Int. 2021. DOI: 10.1155/2021/6546170. DOI: https://doi.org/10.1155/2021/6546170
Sonu, P.; Kishore, M.; Naik, S.J. K.; Sandhya, N.; Pawar, A.C.; Rajeswari, S.; Nagesh, B. Int. J. Pharm. Res. 2022, 14. DOI:10.31838/ijpr/2022.14.01.005. DOI: https://doi.org/10.33545/26647222.2022.v4.i1a.45
Rampogu, S.; Balasubramaniyam, T.; Lee, J.H. Biomed. Pharmacother. 2022, 155, 113760. DOI: 10.1016/j.biopha.2022.113760. DOI: https://doi.org/10.1016/j.biopha.2022.113760
Babbar, N. Pharma Innovation. 2022, 4, 74-75.
Rzhetsky, A.; Nei, M. Mol. Biol. Evol. 1992, 9:945-967. DOI: https://doi.org/10.1093/oxfordjournals.molbev.a040771.
Tamura K.; Stecher G.; Kumar S. MEGA 11. Molecular Evolutionary Genetics Analysis Version 11. 2021. DOI: https://doi.org/10.1093/molbev/msab120.
Felsenstein J. Evolution. 1985, 39:783-791. DOI: https://doi.org/10.2307/2408678. DOI: https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Zuckerkandl, E.; Pauling, L. Academic Press. 1965, pp. 97-166. DOI: https://doi.org/10.1016/B978-1-4832-2734-4.50017-6.
Nei M.; Kumar, S. Molecular Evolution and Phylogenetics, Oxford University Press, New York, 2000. DOI: https://doi.org/10.1093/oso/9780195135848.001.0001
Saitou N.; Nei, M. Mol. Biol. Evol. 1987, 4:406-425.
Marchler-Bauer, A.; Bo, Y.; Han L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R. Nucleic Acids Res. 2017, 45, D200-D203. DOI:10.1093/nar/gkw1129. DOI: https://doi.org/10.1093/nar/gkw1129
Letunic, I.; Bork, P. Nucleic Acids Res. 2018, 46, D493-D496. DOI:10.1093/nar/gkx922. DOI: https://doi.org/10.1093/nar/gkx922
Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A. Nucleic Acids Res. 2016, 44, D279 D285. DOI:10.1093/nar/gkv1344. DOI: https://doi.org/10.1093/nar/gkv1344
Haft, D.H; Selengut J.D.; Richter, R.A.; Harkins, D.; Basu, M.K.; Beck, E. Nucleic Acids Res. 2013, 41, D387-D395. DOI:10.1093/nar/gks1234. DOI: https://doi.org/10.1093/nar/gks1234
Selengut J.D.; Haft, D.H.; Davidsen, T.; Ganapathy, A.; Gwinn-Giglio, M.; Nelson W.C.; Nucleic Acids Res. 2007, 35, D260-264. DOI:10.1093/nar/gkl1043. DOI: https://doi.org/10.1093/nar/gkl1043
Altschul, S.F.; Madden, T.L.; Sch ffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W. Nucleic Acids Res. 1997, 25, 3389 3402. DOI:10.1093/nar/25.17.3389. DOI: https://doi.org/10.1093/nar/25.17.3389
Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K. BLAST+: architecture and applications. BMC Bioinformatics. 2009, 10, 421. DOI:10.1186/1471-2105-10-421. DOI: https://doi.org/10.1186/1471-2105-10-421
Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino-Gallo, T.; Bertoni, M.; Bordoli, L.; Schwede, T. Nucleic Acids Res. 2014, 42, W252-W258. DOI: 10.1093/nar/gku340. DOI: https://doi.org/10.1093/nar/gku340
Wilmouth, R.C.; Turnbull, J.J.; Welford, R.W.; Clifton, I.J.; Prescott, A.G.; Schofield, C.J. Structure. 2002, 10, 93-103. DOI: 10.1016/s0969-2126(01)00695-5. DOI: https://doi.org/10.1016/S0969-2126(01)00695-5
http://www.worldfloraonline.org, accessed in november 2022.
Sharkey, T.D. J. Exp. Bot. 2022, 74, 510–519 DOI: https://doi.org/10.1093/jxb/erac254.
Bracher, A.; Whitney S.M.; Hartl, F. U.; Hayer-Hartl, M. Annu. Rev. Plant. Biol. 2017, 68:29–60. https://doi.org/10.1146/annurev-arplant-043015-111633.
Valegård, K.; Hasse, D.; Andersson, I.; Gunn, L. H. Acta Crystallogr. D. Struct. Biol. 2018, 74, 1–9. DOI: https://doi.org/10.1107/S2059798317017132.
Edwards, E. J.; Donoghue, M.J. Am. Nat. 2006, 167, 777-793. DOI: 10.1086/504605. DOI: https://doi.org/10.1086/504605
Bouvier, J.W.; Emms, D.M.; Rhodes, T.; Bolton, J.S.; Brasnett, A.; Eddershaw, A.; Nielsen, J.R.; Unitt, A.; Whitney, S. M.; Kelly, S. Mol. Biol. Evol. 2021, 38, 2880-2896. DOI: https://doi.org/10.1093/molbev/msab079.
Casola, C.; Li, J. PeerJ. 2022, 10, e12791. DOI: 10.7717/peerj.12791. DOI: https://doi.org/10.7717/peerj.12791
Barrios, D.; González-Torres, L.R.; Arias, S.; Majure, L.C. Plant Syst. Evol. 2020, 306, 63. DOI: https://doi.org/10.1007/s00606-020-01693-5.
Wink, M. Phytochem. 2003, 64, 3-19. DOI:10.1016/S0031-9422(03)00300-5. DOI: https://doi.org/10.1016/S0031-9422(03)00300-5
Rawlings, N.D.; Tolle, D.P.; Barrett, A.J. Biochem. J. 2004, 378, 705-716. DOI: 10.1042/BJ20031825. DOI: https://doi.org/10.1042/bj20031825
Qi, X.; Bakht, S.; Leggett, M.; Maxwell, C.; Melton, R.; Osbourn, A. PNAS. 2004, 101, 8233-8238. DOI: https://doi.org/10.1073/pnas.0401301101.

Downloads
Published
Issue
Section
License
Copyright (c) 2023 Tamayo-Ordoñez Y.J., Tamayo-Ordoñez M.C., Rosas-García N.M., Sosa-Santillán G.J., Ayil-Gutiérrez B.A.

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.