In silico Studies of N-(4-tert-butylphenyl)-4-(3-chloropyridin-2-yl) piperazine-1-carboxamide Derivatives as Potent TRPV1 Antagonists using 3D QSAR, ADMET and Molecular Docking
DOI:
https://doi.org/10.29356/jmcs.v70i1.2105Keywords:
TRPV1antagonist, 3D-QSAR, Molecular docking, ADMET prediction, BCTC derivativesAbstract
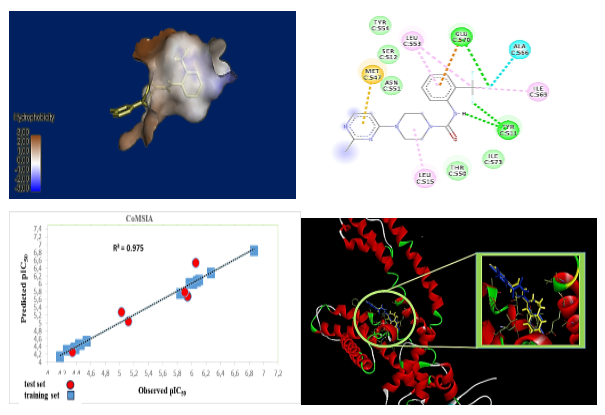
Abstract. TRPV1 is a promising therapeutic target given its involvement in pain management and inflammation.TRPV1 antagonists are increasingly sought after for their analgesic, anti-inflammatory and antitumor properties with fewer side effects. This study focused on the design of new effective TRPV1 antagonists by replacing the pyridine ring of BCTC with a pyrimidine ring. Significant 3D-QSAR models were developed using CoMSIA and CoMFA methods and showed a satisfactory correlation between experimental and predicted activity (Q2 = 0.715; R2 = 0.988; SEE = 0.048). Electrostatic, hydrophobic fields and hydrogen bond acceptors contributed significantly to the biological activity of studied compounds as well as the importance of hydrophobic , electrostatic fields and H-bond acceptors on the antagonistic activity of the most active molecule in the series of compounds studied. Molecular docking analysis validated the 3D-QSAR models and explained the interactions of the most active ligands with the binding site. These results permitted the prediction of new compounds, whose pharmacokinetic properties, toxicity and pharmacodynamics effects were assessed using ADMET and drug similarity.
Resumen. El TRPV1 es un objetivo terapéutico prometedor debido a su participación en el manejo del dolor y la inflamación. Los antagonistas del trpv1 son cada vez más buscados por sus propiedades analgésicas, antiinflamatorias y antitumorales con menos efectos secundarios. Este estudio se centró en el diseño de nuevos antagonistas efectivos del trpv1 mediante la sustitución del anillo de piridina del bctc por un anillo de pirimidina. Se desarrollaron modelos 3D-QSAR significativos utilizando los métodos Comsia y Comfa, que mostraron una correlación satisfactoria entre la actividad experimental y la prevista (Q2 = 0.715; R2 = 0.988; SEE = 0.048). Los campos electrostáticos e hidrofóbicos y los aceptores de enlaces de hidrógeno contribuyeron significativamente a la actividad biológica de los compuestos estudiados. El análisis de acoplamiento molecular validó los modelos 3D-QSAR y explicó las interacciones de los ligandos más activos con el sitio de unión. Estos resultados permitieron predecir nuevos compuestos, cuyas propiedades farmacocinéticas, toxicidad y efectos farmacodinámicos se evaluaron mediante ADMET y por similitud con fármacos.
Downloads
References
1. Smit, T.; Mayorga, N. A.; Rogers, A. H.; Nizio, P.; Zvolensky, M. J. Addict. Behav. 2023, 136, 107495. DOI: https://doi.org/10.1016/j.addbeh.2022.107495.
2. Schrepf, A.; Gallop, R.; Naliboff, B.; Harte, S. E.; Afari, N.; Lai, H. H.; Pontari, M.; McKernan, L. C.; Strachan, E.; Kreder, K. J.; et al. J. Pain 2022, 23, 1594–1603. DOI: https://doi.org/10.1016/j.jpain.2022.03.240.
3. Reilly, R. M.; McDonald, H. A.; Puttfarcken, P. S.; Joshi, S. K.; Lewis, L. G.; Pai, M.; Franklin, P. H.; Segreti, J. A.; Neelands, T. R.; Han, P.; et al. J. Pharmacol. Exp. Ther. 2012, 342, 416–428. DOI: https://doi.org/10.1124/jpet.111.190314.
4. Aghazadeh Tabrizi, M.; Baraldi, P. G.; Baraldi, S.; Gessi, S.; Merighi, S.; Borea, P. A. Med. Res. Rev. 2017, 37, 936–983. DOI: https://doi.org/10.1002/med.21427.
5. Toughzaoui, A.; Chedadi, O.; El Aissouq, A.; El Ouardi, Y.; Bouachrine, M.; Ouammou, A. Phys. Chem. Res. 2022, 11, 353–368. DOI: https://doi.org/10.22036/pcr.2022.334832.2059.
6. Csekő, K.; Beckers, B.; Keszthelyi, D.; Helyes, Z. Pharmaceuticals. 2019, 12, 48. DOI: https://doi.org/10.3390/ph12020048.
7. Seebohm, G.; Schreiber, J. A. Cell. Physiol. Biochem. 2021, 22, 108–130. DOI: https://doi.org/10.33594/000000358.
8. Loyd, D. R.; Chen, P. B.; Hargreaves, K. M. Neuroscience. 2012, 203, 207–215. DOI: https://doi.org/10.1016/j.neuroscience.2011.12.019.
9. Sawhney, J. P.; Kothiwale, V. A.; Bisne, V.; Durgaprasad, R.; Jadhav, P.; Chopda, M.; Vanajakshamma, V.; Meena, R.; Vijayaraghavan, G.; Chawla, K.; et al. Indian Heart J. 2018, 70. DOI: https://doi.org/10.1016/j.ihj.2018.09.001.
10. Helyes, Z.; Csekő, K.; Beckers, B.; Keszthelyi, D.Front. Pharmacol. 2017, 8,121. DOI: https://doi.org/10.3389/fphar.2017.00121.
11. Li, J.; Nie, C.; Qiao, Y.; Hu, J.; Li, Q.; Wang, Q.; Pu, X.; Yan, L.; Qian, H. Eur. J. Med. Chem. 2019, 178, 433–445. DOI: https://doi.org/10.1016/j.ejmech.2019.06.007.
12. Othman, A. A.; Nothaft, W.; Awni, W. M.; Dutta, S. Br. J. Clin. Pharmacol. 2013, 75, 203–212. DOI: https://doi.org/10.1111/j.1365-2125.2012.04405.x.
13. Benko, R.; Illényi, L.; Kelemen, D.; Papp, R.; Papp, A.; Bartho, L. Eur. J. Pharmacol. 2012, 674, 44–50. DOI: https://doi.org/10.1016/j.ejphar.2011.10.021.
14. Abdelgawad, M. A.; Elkanzi, N. A. A.; Musa, A.; Ghoneim, M. M.; Ahmad, W.; Elmowafy, M.; Abdelhaleem Ali, A. M.; Abdelazeem, A. H.; Bukhari, S. N. A.; El-Sherbiny, M.; et al. Arab. J. Chem. 2022, 15, 104015 DOI:.https://doi.org/10.1016/j.arabjc.2022.104015.
15. Khurana, L.; Fu, B.; Duddupudi, A. L.; Liao, H.; Immadi, S. S.; Kendall, D. A.; Lu, D. J. Med. Chem. 2017. DOI: https://doi.org/10.1021/acs.jmedchem.6b01448.
16. Burkert, U.; Allinger, N. L., in: Molecular Mechanics; American Chemical Society: Washington, DC, 1982.
17. Sousa, S. F.; Ribeiro, A. J. M.; Coimbra, J. T. S.; Neves, R. P. P.; Martins, S. A.; Moorthy, N. S. H. N.; Fernandes, P. A.; Ramos, M. J. Curr. Med. Chem. 2013, 20, 2296–2314.DOI: https://doi.org/10.2174/0929867311320180002.
18. Shamshad, H.; Hafiz, A.; Althagafi, I. I.; Saeed, M.; Mirza, A. Z. Curr. Comput.-AidedDrugDes. 2019, 16. DOI: https://doi.org/10.2174/1573409915666190827163327.
19. Nie, C.; Li, Q.; Qiao, Y.; Hu, J.; Gao, M.; Wang, Y.; Qiao, Z.; Wang, Q.; Yan, L.; Qian, H. Eur. J. Med. Chem. 2020, 194, 112236.DOI: https://doi.org/10.1016/j.ejmech.2020.112236.
20. Li, J.; Nie, C.; Qiao, Y.; Hu, J.; Li, Q.; Wang, Q.; Pu, X.; Yan, L.; Qian, H. Eur. J. Med. Chem. 2019, 178, 433–445. DOI: https://doi.org/10.1016/j.ejmech.2019.06.007.
21. Tong, L.; Guo, L.; Lv, X.; Li, Y. J. Mol. Graphics Modell. 2017, 71, 1–12. DOI: https://doi.org/10.1016/j.jmgm.2016.10.012.
22. Clark, M.; Cramer, R. D., III; Opdenbosch, N. V. J. Comput. Chem. 1989, 10, 982–1012. DOI: https://doi.org/10.1002/jcc.540100804.
23. Chedadi, O.; El Aissouq, A.; El Ouardi, Y.; Bouachrine, M.; Ouammou, A. Biointerface Res. Appl. Chem. 2022, 12, 5100–5115. DOI: https://doi.org/10.33263/BRIAC124.51005115.
24. El Aissouq, A.; Chedadi, O.; Bouachrine, M.; Ouammou, A. J. Chem. 2021, 1901484. DOI: https://doi.org/10.1155/2021/1901484.
25. El Aissouq, A.; El Hamdani, S.; El Mchichi, A.; Bouachrine, M. Moroccan J. Chem. 2022, 10. DOI: https://doi.org/10.48317/IMIST.PRSM/morjchem-v10i4.34319.
26. Wang, A.; Yang, Y.; Jun, Y.; Wang, B.; Lv, K.; Liu, M.; Guo, H.; Lu, Y. Bioorg. Med. Chem. 2018, 26, 2073–2084. DOI: https://doi.org/10.1016/j.bmc.2018.03.004.
27. Vrtačnik, M.; Voda, K. Chemosphere 2003, 52, 1689–1699. DOI: https://doi.org/10.1016/S0045-6535(03)00354-0.
28. A. Chiari, L. P.; da Silva, A. P.; Honório, K. M.; da Silva, A. B. F. J. Mol. Model. 2023, 29, 1–3. DOI: https://doi.org/10.1007/s00894-023-05443-5.
29. Ai, Y.; Wang, S. T.; Sun, P. H.; Song, F. J. Int. J. Mol. Sci. 2011, 12, 1605–1624. DOI:https://doi.org/10.3390/ijms12031605.
30. Srivastava, P.; Tripathi, P. N.; Sharma, P.; Rai, S. N.; Singh, S. P.; Srivastava, R. K.; Shankar, S.; Shrivastava, S. K. Eur. J. Med. Chem. 2019, 163, 116–135. DOI: https://doi.org/10.1016/j.ejmech.2018.11.049.
31. Dar, A. M.; Mir, S. J. Anal. Bioanal. Tech. 2017, 8, 8–10. DOI: https://doi.org/10.4172/2155-9872.1000356.
32. El Bahi, S.; Boutalaka, M.; El Alaouy, M. A.; Bouamrane, S.; Alaqarbeh, M.; Choukrad, M.; Sbai, A.; Bouachrine, M.; Lakhlifi, T. New J. Chem. 2023, 47, 12816–12829. DOI: https://doi.org/10.1039/D3NJ02471G.
33. Moukhliss, Y.; Koubi, Y.; Alaqarbeh, M.; Rehman, H. M.; Maghat, H.; Sbai, A.; Bouachrine, M.; Lakhlifi, T. ChemistrySelect 2023, 8, e202203908. DOI: https://doi.org/10.1002/slct.202203908.
34. 34 Madhavilatha, N.; Rama, G.; Babu, M.; Afriza, D.; Suriyah, W. H.; Ichwan, S. J. A. J. Phys.: Conf. Ser. 2018, 1073, 032001. DOI: https://doi.org/10.1088/1742-6596/1073/3/032001.
35. Varadharajan, A.; Sinha, S.; Xu, A.; Daniel, A.; Kim, K.; Shanmugam, N.; Wu, E.; Yang, C.; Zhang, M.; Acree, W. E. J. Solution Chem. 2023, 52, 70–90. DOI: https://doi.org/10.1007/s10953-022-01215-6.
36. El Aissouq, A. E.; Chedadi, O.; Bouachrine, M.; Khalil, F. J. Biomol. Struct. Dyn. 2022, 1,14. DOI: https://doi.org/10.1080/07391102.2022.2071341.
37. Boutalaka, M.; El Bahi, S.; Alaqarbeh, M.; El Alaouy, M. A.; Koubi, Y.; Khatabi, K. E.; Maghat, H.; Bouachrine, M.; Lakhlifi, T. J. Biomol. Struct. Dyn. 2023, 1–20. DOI: https://doi.org/10.1080/07391102.2023.2233629.
38. Voight, E. A.; Gomtsyan, A. R.; Daanen, J. F.; Perner, R. J.; Schmidt, R. G.; Bayburt, E. K.; Didomenico, S.; McDonald, H. A.; Puttfarcken, P. S.; Chen, J.; et al. J. Med. Chem. 2014, 57 (17), 7412–7424. DOI: https://doi.org/10.1021/jm500916t.
39. Reda El-Mernissi Ayoub Khaldan, T. L.; Bouamrane, S.; Rehman, H. M.; Alaqarbeh, M.; Ajana, M. A.; Bouachrine, M. J. Biomol. Struct. Dyn. 2023, 1–18. DOI: https://doi.org/10.1080/07391102.2023.2214233.
40. Roth, H. S.; Botham, R. C.; Schmid, S. C.; Fan, T. M.; Dirikolu, L.; Hergenrother, P. J. J. Med. Chem. 2015, 58, 4046–4065. DOI: https://doi.org/10.1021/acs.jmedchem.5b00413.
41. Daina, A.; Michielin, O.; Zoete, V. Sci. Rep. 2017, 7,42717. DOI: https://doi.org/10.1038/srep42717.
42. Abdessadak, O.; Alaqarbeh, M.; Zaki, H.; Almohtaseb, F.; Alsakhen, N.; Ajana, M. A.; Lakhlifi, T.; Bouachrine, M. Struct. Chem. 2023, 34, 1173–1187. DOI: https://doi.org/10.1007/s11224-022-02068-x.

Downloads
Published
Issue
Section
License
Copyright (c) 2025 Abdelilah Toughzaoui, Oussama Chedadi , Abdellah El Aissouq, Youssef El Ouardi, Mohammed Bouachrine, Abdelkrim Ouammou, Kamal Moradi

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.