Synthesis, Characterization, and Molecular Docking Against a Receptor Protein FimH of Escherichia coli (4XO8) of Thymidine Derivatives
DOI:
https://doi.org/10.29356/jmcs.v65i2.1464Keywords:
Thymidine, synthesis, Escherichia coli, molecular docking, DFT, inhibitor, lectin protein (FimH)Abstract
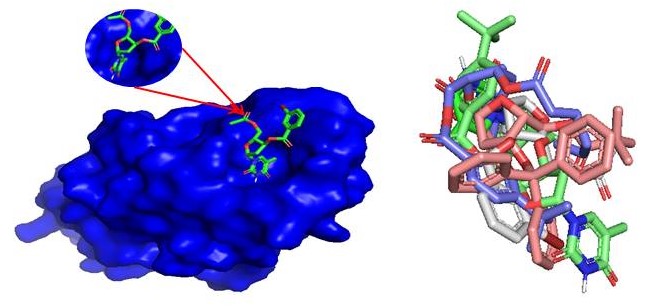
Abstract. Thymidine is known as a progenitor of nucleosides that have significant biological activity. The widening importance of nucleoside derivatives as unrivaled potential antimicrobial and therapeutic agents has attracted contemplation to the synthesis of thymidine derivatives. In the present study, thymidine was treated with various acyl halides to produce 5ʹ-O-acyl thymidine derivatives by direct acylation method with an excellent yield. To obtain newer products for antimicrobial assessment studies, the 5ʹ-O-thymidine derivatives were further modified into three series of 3ʹ-O-acyl thymidine derivatives containing a wide variety of functionalities in a single molecular framework. The chemical structures of the newly synthesized compounds were elucidated by analyzing their physicochemical, elemental, and spectroscopic data. Additionally, the X-ray powder diffraction (XRD) of these acylated products was studied. For the computational investigation, we have selected eight synthesized thymidine derivatives, which have notable antibacterial activity, and performed molecular docking against bacterial lectin protein FimH of Escherichia coli (4XO8) to suggest a potent inhibitor against bacterial function. Molecular docking was performed using AutoDock Vina to calculate the binding affinities and interactions between the antibacterials and the FimH E. coli (4XO8). It was found that the selected thymidine derivatives have strongly interacted mainly with Tyr48, Tyr137, Asp140, Arg98, Gln133, Phe1, Asn23, Asn135, Lys76, Asp47, Ile13, and Ile52 residues. In silico pharmacokinetic properties were also predicted to search their absorption, metabolism, excretion, and toxicity. This computational examination showed that these thymidine derivatives might be used as potential inhibitors against the promising antibacterial activity for future studies.
Resumen. Se prepararon varios derivados 5ʹ-O-acil timidínicos por acilación directa con rendimientos excelentes que fueron transformados en tres series de derivados 3ʹ-O-acil timidínicos con una amplia variedad de funcionalidades. Estos compuestos fueron la base de un estudio de docking dirigido a la lectina bacteriana FimH de Escherichia coli (4XO8) con la finalidad de proponer un inhibidor contra esta función bacteriana.
Downloads
References
Jordheim, L. P.; Durantel, D.; Zoulim, F.; Charles, D. Nat. Rev. Drug Discov. 2013, 12, 447–464.
Galmarini, C. M.; Mackey, J. R.; Dumontet, C. Lancet Oncol. 2002, 3, 415–424. DOI: https://doi.org/10.1016/S1470-2045(02)00788-X
Damaraju, V. L.; Damaraju, S.; Young, J. D.; Stephen, A. B.; John, M.; Michael, B S.; Carol, E. C. Oncogene 2003, 22, 7524–7536. DOI: https://doi.org/10.1038/sj.onc.1206952
Eyer, L.; Nencka, R.; de Clercq, E.; Katherine, S. R.; R?žek D. Antivir. Chem. Chemother. 2018, 26, 1–28. DOI: https://doi.org/10.1177/2040206618761299
Vittori, S.; Ben, D. D.; Lambertucci, C.; Marucci, G.; Volpini, R.; Cristalli, G. Curr. Med. Chem. 2006, 13, 3529–3552. DOI: https://doi.org/10.2174/092986706779026228
Sax, N. I.; Lewis, R. J. The Following List of Chemicals Known or Believed to be Teratogens is Drawn Primarily from Dangerous Properties of Industrial Materials, 7th ed., John Wiley & Sons, Hoboken, N.J. 1988.
Ohrui, H. Proc. Jpn. Acad. Ser. 2011, 87B, 53–65. DOI: https://doi.org/10.2183/pjab.87.53
Jordheim, L. P.; Durantel, D.; Zoulim, F.; Dumontet, C. Nat. Rev. Drug Discov. 2013, 12, 447–464. DOI: https://doi.org/10.1038/nrd4010
Bacon, T. H.; Levin, M. J.; Leary, J. J.; Robert, Sarisky, T.; David, S. Clin. Microbiol. Rev. 2003, 16, 114–128. DOI: https://doi.org/10.1128/CMR.16.1.114-128.2003
Hamuy , R.; Berman, B. Drugs Today (Barc.) 1998, 34, 1013–1025. DOI: https://doi.org/10.1358/dot.1998.34.12.487486
Tabata, S.; Yamamoto, M.; Goto, H.; Akiyoshi, H.; Maki, O.; Takuya, K.; Atsushi, M,; Ryuji, I.; Misako, H.; Kohichi, K.; Yoshinari, S.; Kentaro, M.; Atsuro, S.; Masaki, H.; Yasuhiko, N.; Saburo, S.; Hiroyasu, E.; Masaru, T.; Tomoyoshi, S.; Tatsuhiko, F.; Shin-Ichi, A. Cell Report 2017, 19, 1313–1321. DOI: https://doi.org/10.1016/j.celrep.2017.04.061
Mitsuya, H.; Wetnholdt, K. J.; Furmant, P. A.; Clairt, M. H.; Nusinoff L. S.; Gallos, R. C.; Bologesit, D.; Arryt, D. W.; Broder, S. Proc. Nalt. Acad. Sci. 1985, 82, 7096–7100. DOI: https://doi.org/10.1073/pnas.82.20.7096
Hurst, M.; Noble, S. Drugs 1999, 58, 919–949. DOI: https://doi.org/10.2165/00003495-199958050-00012
Itoh, M.; Hagiwara, D.; Notani, J. Synthesis 1975, 1975, 456–457. DOI: https://doi.org/10.1055/s-1975-23804
Tsuda, Y.; Haque, M. E. Chem. Pharm. Bull. (Japan) 1983, 31, 1437–1439. DOI: https://doi.org/10.1248/cpb.31.1437
Adinolfi, M.; Barone, G.; Iadonisi, A.; Schiattarella, M. Tetrahedron Lett. 2003, 44, 4661–4663. DOI: https://doi.org/10.1016/S0040-4039(03)01072-4
Yan, Y. L.; Guo, J. R.; Liang, C. F. Chem. Asian J. 2017, 12, 2471–2479. DOI: https://doi.org/10.1002/asia.201700867
Ch, R.; Tyagi, M.; Patil, P. R.; Kartha, K. P. R. Tetrahedron Lett. 2011, 52, 5841–5846. DOI: https://doi.org/10.1016/j.tetlet.2011.08.141
Andary, C.; Wylde, R.; Laffite, C.; Privat, G.; Winternitz, F. Phytochemistry 1982, 21, 1123–1127. DOI: https://doi.org/10.1016/S0031-9422(00)82429-2
Ishji, H.; Nakamura, M.; Seo, S.; Tori, K.; Tozyo, T.; Yoshimura, Y. Chem. Pharm. Bull. (Japan) 1980, 28, 2367–2373. DOI: https://doi.org/10.1248/cpb.28.2367
Kabir, A. K. M. S.; Dutta, P.; Anwar, M. N. Chittagong Univ. J. Sci. 2005, 29, 1– 8.
Mirja, H.; Thisbe K. L. Eur. J. Org. Chem. 2011, 20-21, 3583–3609. DOI: https://doi.org/10.1002/ejoc.201100407
Carreras, E.; Boix, E.; Navarro, S.; Rosenberg, H. F.; Cuchillo, C. M.; Nogues, M. V. Mol.Cell. Biochem. 2005, 272, 1–7. DOI: https://doi.org/10.1007/s11010-005-4777-2
Marc, T.; Nogues, M. V.; Boix, E. J. Mol. Recognit. 2011, 24, 90–100. DOI: https://doi.org/10.1002/jmr.1027
Chatfield, D.; Christopher, J. Theor. Chem. Accounts Theor. Comput. Model (Theoretica Chim. Acta) 2002, 108, 367–368. DOI: https://doi.org/10.1007/s00214-002-0380-8
Kawsar, S. M. A.; Hamida, A. A.; Sheikh, A. U.; Hossain, M. K.; Shagir, A. C.; Sanaullah, A. F. M.; Manchur, M. A.; Imtiaj, H.; Ogawa, Y.; Fujii, Y.; Koide, Y.; Ozeki, Y. Int. J. Org. Chem. 2015, 5, 232–245. DOI: https://doi.org/10.4236/ijoc.2015.54023
Shagir, A. C.; Bhuiyan, M. M. R.; Ozeki, Y.; Kawsar, S. M. A. Curr. Chem. Lett. 2016, 5, 83–92.
Said, S. A. J.; Anwar, U. H.; Abdul, R. I.; Mohammed, S. S. Powder Technol. 2007, 175, 115–121. DOI: https://doi.org/10.1016/j.powtec.2007.01.013
Gaussian09, R. A.; Frisch, M. J.; Trucks, G. W.; Schlegel, H. B.; Scuseria, G. E.; Robb, M. A.; Cheeseman, J. R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G. A.; et al. Gaussian, Inc, Wallingford CT. 2009.
Becke, A. D. Phys. Rev. 1988, 38A, 3098–3100. DOI: https://doi.org/10.1103/PhysRevA.38.3098
Lee, C.; Yang, W.; Parr, R. G. Phys. Rev. 1988, 37B, 785–789. DOI: https://doi.org/10.1103/PhysRevB.37.785
Berman, H. M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T. N.; Weissig, H. Nucleic Acids Res. 2000, 28, 235–242. DOI: https://doi.org/10.1093/nar/28.1.235
Delano, W. L. The PyMOL molecular graphics system, San Carlos, CA, USA, 2002.
Guex, N.; Peitsch, M. C. Electrophoresis, 1997, 18, 2714–2723. DOI: https://doi.org/10.1002/elps.1150181505
Dallakyan, S.; Olson, A. J. Small-Molecule Library Screening by Docking with Py Rx. In: Hempel, J. E.; Williams, C. H.; Hong, C. C. (eds.). Chemical Biology: Methods and Protocols, Springer, New York, USA, 2015, 243-250. DOI: https://doi.org/10.1007/978-1-4939-2269-7_19
Version ADS 4.0, Accelrys, San Diego, USA, 2017.
Onodera, K.; Satou, K.; Hirota, H. J. Chem. Inf. Model. 2007, 47, 1609–1618. DOI: https://doi.org/10.1021/ci7000378
Warren, G. L.; Andrews, C. W.; Capelli, A. M.; Clarke, B.; LaLonde, J.; Lambert, M. H.; Lindvall, M.; Nevins, N.; Semus, S. F.; Senger, S.; Tedesco, G.; Wall, I. D.; Woolven, J. M.; Peishoff, C. E.; Head, M. S. J. Med. Chem. 2006, 49, 5912–5931. DOI: https://doi.org/10.1021/jm050362n
Ferreira, L. L. G.; Andricopulo, A. D. Drug Discov. Today 2019, 24, 1157–1165. DOI: https://doi.org/10.1016/j.drudis.2019.03.015
Devi, S. R.; Jesmin, S.; Rahman, M.; Manchur, M. A.; Fujii, Y.; Kanaly, R. A.; Ozeki, Y.; Kawsar, S. M. A. ACTA Pharm. Sci. 2019, 57, 47–68. DOI: https://doi.org/10.23893/1307-2080.APS.05704
Jesmin, S.; Devi, S. R.; Rahman, M.; Islam, M.; Kanaly, R. A.; Fuji, Y.; Hayashi, N.; Ozeki, Y.; Kawsar, S. M. A. J. Bang. Chem. Soc. 2017, 29, 12–20.
Arifuzzaman, M.; Mirajul, M. I.; Monjur, M. R.; Atiar, M. R.; Kawsar, S. M. A. ACTA Pharm. Sci. 2018, 56, 7–22. DOI: https://doi.org/10.23893/1307-2080.APS.05622
Kroemer, R. T.; Vulpetti, A.; McDonald, J. J.; Rohrer, D. C.; Trosset, J. Y.; Giordanetto, F.; Cotesta, S.; McMartin, C.; Kihlen, M.; Stouten, P. F. W. J. Chem. Inf. Comput. Sci. 2004, 44, 871–881. DOI: https://doi.org/10.1021/ci049970m
Jones, G.; Willett, P.; Glen, R. C.; Leach, A. R.; Taylor, R. J. Mol. Biol. 1997, 267, 727–748. DOI: https://doi.org/10.1006/jmbi.1996.0897
Kontoyianni, M.; McClellan, L. M.; Sokol, G. S. J. Med. Chem. 2004, 47, 558–565. DOI: https://doi.org/10.1021/jm0302997
Perlstein, J. J. Am. Chem. Soci. 2001, 123, 191–192.
Pires, D. E. V.; Blundell, T. L.; Ascher, D. B. J. Med. Chem. 2015, 58, 4066–4072. DOI: https://doi.org/10.1021/acs.jmedchem.5b00104
Clark, D. E. Drug Discov. Today 2003, 8, 927–933. DOI: https://doi.org/10.1016/S1359-6446(03)02827-7
Kok-Yong, S.; Lawrence, L. Drug Distribution and Drug Elimination. Basic Pharmacokinetic Concepts and Some Clinical Applications, InTechOpen, London, SW7 2QJ, UK, 2015. DOI: https://doi.org/10.5772/59929
Thapar, M. M.; Pharmacokinetics and Dynamics of Atovaquone and Proguanil, Malarone®, Karolinska University Press, Stockholm, Sweden, 2004.

Downloads
Additional Files
Published
Issue
Section
License
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.