Investigation of Structure, Reactivity, and Biological Activity of Thiazole-Containing Compounds: A Computational Study
DOI:
https://doi.org/10.29356/jmcs.v70i1.2378Keywords:
Thiazole, density functional theory, molecular docking, anticancer, anticholinesterase reactivityAbstract
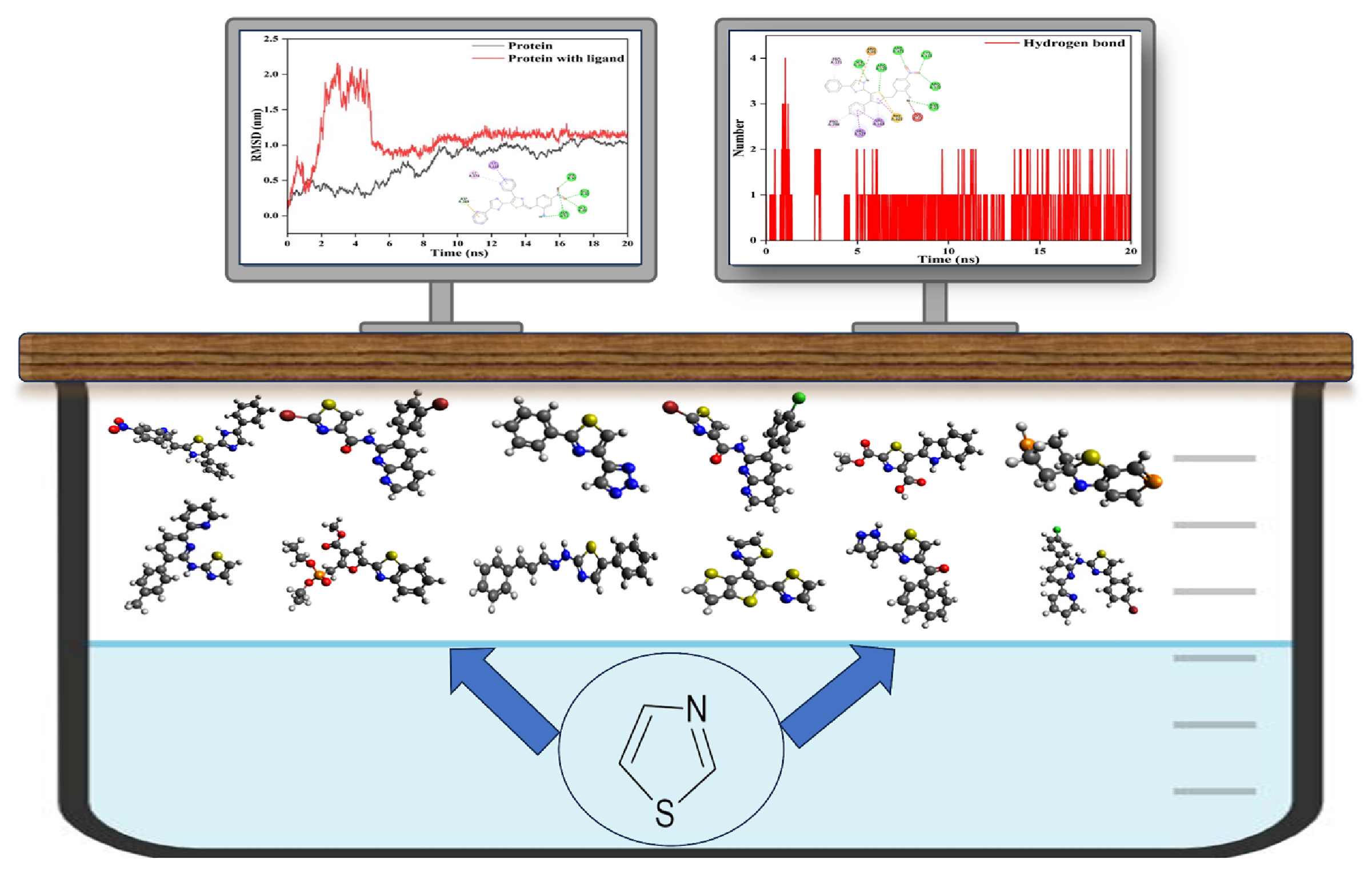
Abstract. Herein, the structure, stability, reactivity, and biological activity of recently synthesized thiazole-containing compounds are evaluated using density functional theory (DFT), molecular docking, and molecular dynamics (MD) techniques. All the selected thiazole-containing compounds are optimized using DFT (B3LYP/def2-TZVPP) method. The DFT reactivity parameters such as energy gap, chemical hardness, chemical potential, ionization potential, electron affinity, electronegativity, softness, and electrophilicity index are calculated. Our calculations indicate that the thiazole-containing compound M1 shows significant structural stability and reactivity. Our physicochemical and pharmacokinetic studies suggest that the selected thiazole-containing compounds possess a drug-like nature. The antibacterial, anticancer, anticholinergic, and antifungal activity of the selected thiazole-containing compounds are investigated using molecular docking and dynamics methods. Our docking studies revealed that M1 shows higher binding affinity with the selected protein targets, which confirms their biological activity. Similarly, M6, M9, and M10 possess lesser binding energy among the selected thiazole-containing compounds. Our MD simulations show that the ligand M1 strongly interacts with the 1M17 protein. shows higher binding affinity with the selected protein targets, which confirms their biological activity. Similarly, M6, M9, and M10 possess lesser binding energy among the selected thiazole-containing compounds. Our MD simulations show that the ligand M1 strongly interacts with the 1M17 protein. This is further evidence that the ligand M1 is a promising candidate for the development of new drugs against deadly pathogens.
Resumen. En este trabajo, se evalúan la estructura, estabilidad, reactividad y actividad biológica de compuestos que contienen tiazol recientemente sintetizados mediante la teoría del funcional de la densidad (DFT), acoplamiento molecular y técnicas de dinámica molecular (MD). Todos los compuestos seleccionados que contienen tiazol se optimizan mediante el método DFT B3LYP/def2-TZVPP. Se calculan los parámetros de reactividad de la DFT, como la brecha de energía, la dureza química, el potencial químico, el potencial de ionización, la afinidad electrónica, la electronegatividad, la suavidad y el índice de electrofilicidad. Nuestros cálculos indican que el compuesto M1, que contiene tiazol, muestra una estabilidad estructural y una reactividad significativas. Nuestros estudios fisicoquímicos y farmacocinéticos sugieren que los compuestos que contienen tiazol seleccionados poseen una naturaleza similar a la de un fármaco. Se investiga la actividad antibacteriana, anticancerígena, anticolinérgica y antifúngica de los compuestos que contienen tiazol seleccionados mediante métodos de acoplamiento molecular y de dinámica. Nuestros estudios de acoplamiento revelaron que M1 presenta una mayor afinidad de unión con las proteínas diana seleccionadas, lo que confirma su actividad biológica. De igual manera, M6, M9 y M10 poseen una menor energía de unión entre los compuestos que contienen los tiazoles seleccionados. Nuestras simulaciones muestran que el ligando M1 interactúa de forma fuerte con la proteína 1M17. Esto constituye una prueba más de que el ligando M1 es un candidato prometedor para el desarrollo de nuevos fármacos contra patógenos mortales.
Downloads
References
1. Tripathi, A. C.; Gupta, S. J.; Fatima, G. N.; Sonar, P. K.; Verma, A.; Saraf, S. K. Eur. J. Med. Chem. 2014, 72, 52–77. DOI: https://doi.org/https://doi.org/10.1016/j.ejmech.2013.11.017
2. Kaminskyy, D.; Kryshchyshyn, A.; Lesyk, R. Eur. J. Med. Chem. 2017, 140, 542–594. DOI: https://doi.org/https://doi.org/10.1016/j.ejmech.2017.09.031
3. Kamble, R. D.; Meshram, R. J.; Hese, S. V.; More, R. A.; Kamble, S. S.; Gacche, R. N.; Dawane, B. S. Comput. Biol. Chem. 2016, 61, 86–96. DOI: https://doi.org/https://doi.org/10.1016/j.compbiolchem.2016.01.007
4. Mohareb, R.; Al-Omran, F.; Abdelaziz, M.; Ibrahim, R. Acta Chim. Slov. 2017, 64, 349–364.
5. Gümüş, M.; Yakan, M.; Koca, İ. Future Med. Chem. 2019, 11, 1979–1998. DOI: https://doi.org/10.4155/fmc-2018-0196
6. Bikobo, D. S. N.; Vodnar, D. C.; Stana, A.; Tiperciuc, B.; Nastasă, C.; Douchet, M.; Oniga, O. J. Saudi Chem. Soc. 2017, 21, 861–868. DOI: https://doi.org/https://doi.org/10.1016/j.jscs.2017.04.007
7. Althagafi, I.; El-Metwaly, N.; Farghaly, T. A. Molecules. 2019, 24, 1741. DOI: https://doi.org/10.3390/molecules24091741
8. Biernasiuk, A.; Kawczyńska, M.; Berecka-Rycerz, A.; Rosada, B.; Gumieniczek, A.; Malm, A.; Dzitko, K.; Łączkowski, K. Z. Med. Chem. Res. 2019, 28, 2023–2036. DOI: https://doi.org/10.1007/s00044-019-02433-2
9. Bondock, S.; Fouda, A. M. Synth. Commun. 2018, 48, 561–573. DOI: https://doi.org/10.1080/00397911.2017.1412465
10. Borcea, A. M.; Ionuț, I.; Crișan, O.; Oniga, O. Molecules. 2021, 26, 624. DOI: https://doi.org/10.3390/molecules26030624
11. Carbone, A.; Cascioferro, S.; Parrino, B.; Carbone, D.; Pecoraro, C.; Schillaci, D.; Cusimano, M. G.; Cirrincione, G.; Diana, P. Molecules. 2021, 26, 81. DOI: https://doi.org/10.3390/molecules26010081
12. Ayati, A.; Emami, S.; Moghimi, S.; Foroumadi, A. Future Med. Chem. 2019, 11, 1929–1952. DOI: https://doi.org/10.4155/fmc-2018-0416
13. Carbone, D.; Vestuto, V.; Ferraro, M. R.; Ciaglia, T.; Pecoraro, C.; Sommella, E.; Cascioferro, S.; Salviati, E.; Novi, S.; Tecce, M. F.; Amodio, G.; Iraci, N.; Cirrincione, G.; Campiglia, P.; Diana, P.; Bertamino, A.; Parrino, B.; Ostacolo, C. Eur. J. Med. Chem. 2022, 234, 114233. DOI: https://doi.org/10.1016/j.ejmech.2022.114233
14. Di Franco, S.; Parrino, B.; Gaggianesi, M.; Pantina, V. D.; Bianca, P.; Nicotra, A.; Mangiapane, L. R.; Lo Iacono, M.; Ganduscio, G.; Veschi, V.; Brancato, O. R.; Glaviano, A.; Turdo, A.; Pillitteri, I.; Colarossi, L.; Cascioferro, S.; Carbone, D.; Pecoraro, C.; Fiori, M. E.; De Maria, R.; Todaro, M.; Screpanti, I.; Cirrincione, G.; Diana, P.; Stassi, G. iScience. 2021, 24, 102664. DOI: https://doi.org/10.1016/j.isci.2021.102664
15. Muhammad, Z. A.; Masaret, G. S.; Amin, M. M.; Abdallah, M. A.; Farghaly, T. A. Med. Chem. 2017, 13, 226–238. DOI: http://dx.doi.org/10.2174/1573406412666160920091146
16. Kryshchyshyn, A.; Roman, O.; Lozynskyi, A.; Lesyk, R. Sci. Pharm. 2018, 86, 26. DOI: https://doi.org/10.3390/scipharm86020026
17. Petrou, A.; Eleftheriou, P.; Geronikaki, A.; Akrivou, M. G.; Vizirianakis, I. Molecules. 2019, 24, 3821. DOI: https://doi.org/10.3390/molecules24213821
18. Khatik, G. L.; Datusalia, A. K.; Ahsan, W.; Kaur, P.; Vyas, M.; Mittal, A.; Nayak, S. K. Curr. Drug Discovery Technol. 2018, 15, 163–177. DOI: https://doi.org/10.2174/1570163814666170915134018
19. Grozav, A.; Porumb, I. D.; Gǎinǎ, L. I.; Filip, L.; Hanganu, D. Molecules 2017, 22, 260. DOI: https://doi.org/10.3390/molecules22020260
20. Djukic, M.; Fesatidou, M.; Xenikakis, I.; Geronikaki, A.; Angelova, V. T.; Savic, V.; Pasic, M.; Krilovic, B.; Djukic, D.; Gobeljic, B.; Pavlica, M.; Djuric, A.; Stanojevic, I.; Vojvodic, D.; Saso, L. Chem.-Biol. Interact. 2018, 286, 119–131. DOI: https://doi.org/10.1016/j.cbi.2018.03.013
21. Liaras, K.; Fesatidou, M.; Geronikaki, A. Molecules. 2018, 23, 685. DOI: https://doi.org/10.3390/molecules23030685
22. Jacob, P. J.; Manju, S. L. Bioorg. Chem. 2020, 100, 103882. DOI: https://doi.org/10.1016/j.bioorg.2020.103882
23. Brito, C. C. B.; da Silva, H. V. C.; Brondani, D. J.; de Faria, A. R.; Ximenes, R. M.; da Silva, I. M.; de Albuquerque, J. F. C.; Castilho, M. S. J. Enzyme Inhib. Med. Chem. 2019, 34, 573–584. DOI: https://doi.org/10.1080/14756366.2018.1550752
24. Rodrigues, C. A.; dos Santos, P. F.; da Costa, M. O. L.; Pavani, T. F. A.; Xander, P.; Geraldo, M. M.; Mengarda, A.; de Moraes, J.; Rando, D. G. G. J. Venomous Anim. Toxins Incl. Trop. Dis. 2018, 24, 31. DOI: https://doi.org/10.1186/s40409-018-0163-x
25. Rouf, A.; Tanyeli, C. Eur. J. Med. Chem. 2015, 97, 911–927. DOI: https://doi.org/10.1016/j.ejmech.2014.10.058
26. Hadziyannis, S. J.; Papatheodoridis, G. V. Expert Rev. Anti-Infect. Ther. 2004, 2, 475–483.
27. Pasqualotto, A. C.; Thiele, K. O.; Goldani, L. Z. Curr. Opin. Invest. Drugs. 2010, 11, 165–174.
28. Kardos, N.; Demain, A. L. Appl. Microbiol. Biotechnol. 2011, 92, 677–687. DOI: https://doi.org/10.1007/s00253-011-3587-6
29. Borelli, C.; Schaller, M.; Niewerth, M.; Nocker, K.; Baasner, B.; Berg, D.; Tiemann, R.; Tietjen, K.; Fugmann, B.; Lang-Fugmann, S.; Korting, H. C. Chemotherapy. 2008, 54, 245–259. DOI: https://doi.org/10.1159/000142334
30. Thierbach, G.; Reichenbach, H. Antimicrob. Agents Chemother. 1981, 19, 504–507. DOI: https://doi.org/10.1128/AAC.19.4.504
31. Li, X. H.; Yang, X. L.; Ling, Y.; Fan, Z. J.; Liang, X. M.; Wang, D. Q.; Chen, F. H.; Li, Z. M. J. Agric. Food Chem. 2005, 53, 2202–2206. DOI: https://doi.org/10.1021/jf0403944
32. Murray, C. J.; Ikuta, K. S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; Johnson, S. C.; Browne, A. J.; Chipeta, M. G.; Fell, F.; Hackett, S.; Haines-Woodhouse, G.; Kashef Hamadani, B. H.; Kumaran, E. A. P.; McManigal, B.; Agarwal, R.; Akech, S.; Albertson, S.; Amuasi, J.; Andrews, J.; Aravkin, A.; Ashley, E.; Bailey, F.; Baker, S.; Basnyat, B.; Bekker, A.; Bender, R.; Bethou, A.; Bielicki, J.; Boonkasidecha, S.; Bukosia, J.; Carvalheiro, C.; Castañeda-Orjuela, C.; Chansamouth, V.; Chaurasia, S.; Chiurchiù, S.; Chowdhury, F.; Cook, A. J.; Cooper, B.; Cressey, T. R.; Criollo-Mora, E.; Cunningham, M.; Darboe, S.; Day, N. P. J.; De Luca, M.; Dokova, K.; Dramowski, A.; Dunachie, S. J.; Eckmanns, T.; Eibach, D.; Emami, A.; Feasey, N.; Fisher-Pearson, N.; Forrest, K.; Garrett, D.; Gastmeier, A.; Giref, A. Z.; Greer, R. C.; Gupta, V.; Haller, S.; Haselbeck, A.; Hay, S. I.; Holm, M.; Hopkins, S.; Iregbu, K. C.; Jacobs, J.; Jarovsky, D.; Javanmardi, F.; Khorana, M.; Kissoon, N.; Kobeissi, E.; Kostyanev, T.; Krapp, F.; Krumkamp, R.; Kumar, A.; Kyu, H. H.; Lim, C.; Limmathurotsakul, D.; Loftus, M. J.; Lunn, M.; Ma, J.; Mturi, N.; Munera-Huertas, T.; Musicha, P.; Mussi-Pinhata, M. M.; Nakamura, T.; Nanavati, R.; Nangia, S.; Newton, P.; Ngoun, C.; Novotney, A.; Nwakanma, D.; Obiero, C. W.; Olivas-Martinez, A.; Olliaro, P.; Ooko, E.; Ortiz-Brizuela, E.; Peleg, A. Y.; Perrone, C.; Plakkal, N.; Ponce-de-Leon, A.; Raad, M.; Ramdin, T.; Riddell, A.; Roberts, T.; Robotham, J. V.; Roca, A.; Rudd, K. E.; Russell, N.; Schnall, J.; Scott, J. A. G.; Shivamallappa, M.; Sifuentes-Osornio, J.; Steenkeste, N.; Stewardson, A. J.; Stoeva, T.; Tasak, N.; Thaiprakong, A.; Thwaites, G.; Turner, C.; Turner, P.; van Doorn, H. R.; Velaphi, S.; Vongpradith, A.; Vu, H.; Walsh, T.; Waner, S.; Wangrangsimakul, T.; Wozniak, T.; Zheng, P.; Sartorius, B.; Lopez, A. D.; Stergachis, A.; Moore, C.; Dolecek, C.; Naghavi, M. Lancet. 2022, 399, 629–655. DOI: https://doi.org/10.1016/S0140-6736(21)02724-0
33. Jonas, O. B.; Irwin, A.; Berthe, F. C. J.; Le Gall, F. G.; Marquez, P. V. World Bank Rep. 2017, 2.
34. Nikalje, A. P. G.; Tiwari, S. V.; Sarkate, A. P.; Karnik, K. S. Med. Chem. Res. 2018, 27, 157–169. DOI: https://doi.org/10.1007/s00044-017-2085-5
35. Dekate, S. M.; Hatzade, K. M.; Ghatole, A. M. Iran. J. Sci. 2023, 47, 953–961. DOI: https://doi.org/10.1007/s40995-023-01496-6
36. Grob, S. Molinspiration Cheminformatics Free Web Services. 2022. https://www.molinspiration.com/, accessed in May 2024.
37. Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B. A.; Thiessen, P. A.; Yu, B.; Zaslavsky, L.; Zhang, J.; Bolton, E. E. Nucleic Acids Res. 2023, 51, D1373–D1380. DOI: https://doi.org/10.1093/nar/gkac956
38. Merecz-Sadowska, A.; Isca, V. M. S.; Sitarek, P.; Kowalczyk, T.; Małecka, M.; Zajdel, K.; Zielińska-Bliźniewska, H.; Jęcek, M.; Rijo, P.; Zajdel, R. Int. J. Mol. Sci. 2024, 25, 4529. DOI: https://doi.org/10.3390/ijms25084529
39. Alecu, I. M.; Zheng, J.; Zhao, Y.; Truhlar, D. G. J. Chem. Theory Comput. 2010, 6, 2872–2887. DOI: https://doi.org/10.1021/ct100326h
40. Neese, F. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2012, 2, 73–78. DOI: https://doi.org/10.1002/wcms.81
41. Dassault Systèmes BIOVIA. Discovery Studio Modeling Environment. Release 2017, San Diego: Dassault Systèmes, 2017.
42. Dallakyan, S.; Olson, A. J. Methods Mol. Biol. 2015, 1263, 243–250. DOI: https://doi.org/10.1007/978-1-4939-2269-7_19
43. Abraham, M. J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J. C.; Hess, B.; Lindahl, E. SoftwareX 2015, 1–2, 19–25.
44. Huang, J.; MacKerell, A. D., Jr. J. Comput. Chem. 2013, 34, 2135–2145. DOI: https://doi.org/10.1002/jcc.23354
45. Cuendet, M. A.; Van Gunsteren, W. F. J. Chem. Phys. 2007, 127, 184102. DOI: https://doi.org/10.1063/1.2779878
46. Uddin, K. M.; Sakib, M.; Siraji, S.; Uddin, R.; Rahman, S.; Alodhayb, A.; Alibrahim, K. A.; Kumer, A.; Matin, M. M.; Bhuiyan, M. M. H. ACS Omega. 2023, 8, 20247–20256. DOI: https://doi.org/10.1021/acsomega.3c01123
47. Trabelsi, S.; Issaoui, N.; Brandán, S. A.; Bardak, F.; Roisnel, T.; Atac, A.; Marouani, H. J. Mol. Struct. 2019, 1185, 223–238. DOI: https://doi.org/10.1016/j.molstruc.2019.02.106
48. Petrou, A.; Fesatidou, M.; Geronikaki, A. Molecules. 2021, 26, 3166. DOI: https://doi.org/10.3390/molecules26113166
49. Shah, P.; Westwell, A. D. J. Enzyme Inhib. Med. Chem. 2007, 22, 527–540. DOI: https://doi.org/10.1080/14756360701425014
50. Mermer, A.; Bayrak, H.; Alyar, S.; Alagumuthu, M. J. Mol. Struct. 2020, 1208, 127891. DOI: https://doi.org/10.1016/j.molstruc.2020.127891
51. Zhou, Z.; Parr, R. G. J. Am. Chem. Soc. 1990, 112, 5720–5724.DOI: https://doi.org/10.1021/ja00171a007
52. Sharom, F. J. Essays Biochem. 2011, 50, 161–178. DOI: https://doi.org/10.1042/BSE0500161
53. Veber, D. F.; Johnson, S. R.; Cheng, H.-Y.; Smith, B. R.; Ward, K. W.; Kopple, K. D. J. Med. Chem. 2002, 45, 2615–2623. DOI: https://doi.org/10.1021/jm020017n
54. Shadrack, D. M.; Ndesendo, V. M. K. Comput. Mol. Biosci. 2017, 7, 1–18. DOI: https://doi.org/10.4236/cmb.2017.71001
55. Kartsev, V.; Geronikaki, A.; Zubenko, A.; Petrou, A.; Ivanov, M.; Glamočlija, J.; Sokovic, M.; Divaeva, L.; Morkovnik, A.; Klimenko, A. Antibiotics. 2022, 11, 1337. DOI: https://doi.org/10.3390/antibiotics11101337
56. Babajan, B.; Anuradha, C. M.; Chaitanya, M.; Gowsia, D.; Kumar, C. S. Int. J. Integr. Biol. 2009, 6, 172–176.
57. Benson, T. E.; Walsh, C. T.; Massey, V. Biochemistry. 1997, 36, 796–805. DOI: https://doi.org/10.1021/bi962220o
58. Frlan, R.; Hrast, M.; Gobec, S. ACS Omega. 2023, 8, 36562–36570. DOI: https://doi.org/10.1021/acsomega.3c04813
59. Jain, P.; Guin, M.; De, A.; Singh, M. J. Phys. Org. Chem. 2023, 36, e4384. DOI: https://doi.org/10.1002/poc.4384
60. De, A.; Ray, H. P.; Jain, P.; Kaur, H.; Singh, N. J. Mol. Struct. 2020, 1199, 126901. DOI: https://doi.org/10.1016/J.MOLSTRUC.2019.126901
61. Mughal, E. U.; Sadiq, A.; Ashraf, J.; Zafar, M. N.; Sumrra, S. H.; Tariq, R.; Mumtaz, A.; Javid, A.; Khan, B. A.; Ali, A.; Javed, C. O. Bioorg. Chem. 2019, 91, 103124. DOI: https://doi.org/10.1016/J.BIOORG.2019.103124
62. Kolandaivel, P.; Selvarengan, P.; Gunavathy, K. V. Biochim. Biophys. Acta, Proteins Proteomics 2006, 1764, 138–145. DOI: https://doi.org/10.1016/j.bbapap.2005.10.016

Downloads
Published
Issue
Section
License
Copyright (c) 2025 Selvarengan Paranthaman, Saranya Gurumoorthy, Karthikeyan Asokan, Abiram Angamuthu

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.