Potential Compounds Interacting in a Specific Potential Site in SARS-CoV-2 Variants, Selected by Molecular Docking
DOI:
https://doi.org/10.29356/jmcs.v66i4.1805Keywords:
S-protein, RBD, COVID-19, SARS-CoV-2 variantsAbstract
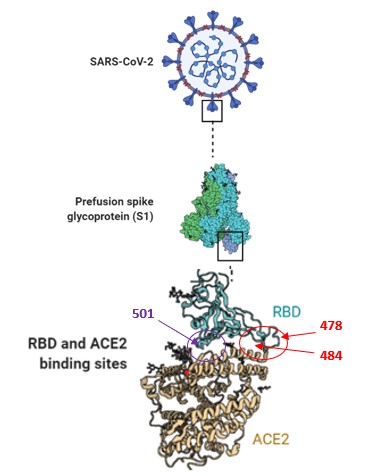
Abstract. The SARS-CoV-2 virus continues developing variants, and different ways of treatments have been proposed during this COVID-19 pandemic. This study proposes compounds to develop a drug against SARS-CoV-2 variants, by molecular docking using a library of compounds (502530 compounds) directed to interact in the region between the amino acids (Ser477, Lys478, Pro479, Cys480, Asn481, Gly482, Val483, Lys484, Gly485, Phe486, Asn487, Cys488, and Tyr489) in the RBD in S-Protein of SARS-CoV-2, this is a specific potential site in SARS-CoV-2 variants.
We propose ten compounds selected by molecular docking, with a high probability to interact in the specific region in the RBD of SARS-CoV-2 variants (amino acids between 478 and 484), to reduce the interaction between S-protein and ACE2. Also, these compounds have a high probability to be safe in humans, validated by web servers of prediction of ADME and toxicity (PreADMET) to develop a new specific adjuvant antiviral against SARS-CoV-2 variants.
Resumen. El virus SARS-CoV-2 continúa desarrollando variantes y se han propuesto diferentes formas de tratamiento durante esta pandemia de COVID-19. Este estudio propone compuestos para desarrollar un fármaco contra las variantes del SARS-CoV-2, mediante simulaciones de acoplamiento molecular (docking) utilizando una quimioteca de compuestos (502530 compuestos) dirigidos a interactuar en la región entre los aminoácidos (Ser477, Lys478, Pro479, Cys480, Asn481, Gly482, Val483, Lys484, Gly485, Phe486, Asn487, Cys488 y Tyr489) en la RBD en la proteína S del SARS-CoV-2, este es un sitio potencial específico en las variantes del SARS-CoV-2.
Proponemos diez compuestos seleccionados por docking, con una alta probabilidad de interactuar en la región específica en la RBD de las variantes del SARS-CoV-2 (aminoácidos entre 478 y 484), para reducir la interacción entre la proteína S y ACE2. Además, estos compuestos tienen una alta probabilidad de ser seguros en humanos, validados por servidores web de predicción de ADME y toxicidad (PreADMET) para desarrollar un nuevo antiviral adyuvante específico contra variantes del SARS-CoV-2.
Downloads
References
University, J.H. COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). https://coronavirus.jhu.edu/map.html, accessed in July 2022.
Conti, P.; Caraffa, A.; Gallenga, C.E.; Kritas, S. K.; Frydas, I.; Younes, A.; Di Emidio, P.; Tetè, G.; Pregliasco, F.; Ronconi, G. J. Biol. Regul. Homeost. Agents. 2021, 35.
Santos, J. C.; Passos, G. A. bioRxiv. 2021. DOI: 10.1101/2020.12.29.424708. DOI: https://doi.org/10.1101/2020.12.29.424708
Luan, B.; Wang, H.; Huynh, T. bioRxiv. 2021. DOI: 10.1101/2021.01.04.425316. DOI: https://doi.org/10.1101/2021.01.04.425316
Aljindan, R.Y.; Al-Subaie, A.M.; Al-Ohali, A.I.; Kumar D, T.; Doss C, G.P.; Kamaraj, B. Comput. Biol. Med. 2021, 135, 104654. DOI: 10.1016/j.compbiomed.2021.104654. DOI: https://doi.org/10.1016/j.compbiomed.2021.104654
Focosi, D.; Maggi, F.; Franchini, M.; McConnell, S.; Casadevall, A. Int. J. Mol. Sci. 2021, 23, 29. DOI: 10.3390/ijms23010029.
Verma, J.; Subbarao, N. Virology. 2021, 561, 107–116. DOI: 10.1016/j.virol.2021.06.009. DOI: https://doi.org/10.1016/j.virol.2021.06.009
Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Cell Res. 2020, 30, 269–271. DOI: 10.1038/s41422-020-0282-0. DOI: https://doi.org/10.1038/s41422-020-0282-0
Sheahan, T. P.; Sims, A. C.; Leist, S. R.; Schäfer, A.; Won, J.; Brown, A. J.; Montgomery, S. A.; Hogg, A.; Babusis, D.; Clarke, M. O.; et al. Nat. Commun. 2020, 11, 222. DOI: 10.1038/s41467-019-13940-6. DOI: https://doi.org/10.1038/s41467-019-13940-6
Li, G.; De Clercq, E. Nat. Rev. Drug Discov. 2020, 19, 149–150. DOI: 10.1038/d41573-020-00016-0. DOI: https://doi.org/10.1038/d41573-020-00016-0
Iftikhar, H.; Ali, H. N.; Farooq, S.; Naveed, H.; Shahzad-ul-Hussan, S. Comput. Biol. Med. 2020, 122, 103848. DOI: 10.1016/j.compbiomed.2020.103848. DOI: https://doi.org/10.1016/j.compbiomed.2020.103848
Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X.; et al. Acta Pharm. Sin. B. 2020, 10, 766–788. DOI: 10.1016/j.apsb.2020.02.008. DOI: https://doi.org/10.1016/j.apsb.2020.02.008
Xia, S.; Yan, L.; Xu, W.; Agrawal, A.S.; Algaissi, A.; Tseng, C.-T. K.; Wang, Q.; Du, L.; Tan, W.; Wilson, I.A.; et al. Sci. Adv. 2019, 5, eaav4580. DOI: 10.1126/sciadv.aav4580. DOI: https://doi.org/10.1126/sciadv.aav4580
Xia, S.; Liu, M.; Wang, C.; Xu, W.; Lan, Q.; Feng, S.; Qi, F.; Bao, L.; Du, L.; Liu, S.; et al. Cell Res. 2020, 30, 343–355. DOI: 10.1038/s41422-020-0305-x. DOI: https://doi.org/10.1038/s41422-020-0305-x
Calligari, P.; Bobone, S.; Ricci, G.; Bocedi, A. Viruses. 2020, 12, 445. DOI: 10.3390/v12040445. DOI: https://doi.org/10.3390/v12040445
Huang, J.; Song, W.; Huang, H.; Sun, Q. J. Clin. Med. 2020, 9, 1131. DOI: 10.3390/jcm9041131. DOI: https://doi.org/10.3390/jcm9041131
Liu, C.; Zhou, Q.; Li, Y.; Garner, L. V.; Watkins, S.P.; Carter, L. J.; Smoot, J.; Gregg, A. C.; Daniels, A. D.; Jervey, S.; et al. ACS Cent. Sci. 2020, 6, 315–331. DOI: 10.1021/acscentsci.0c00272. DOI: https://doi.org/10.1021/acscentsci.0c00272
Locht, C. Anaesth. Crit. Care Pain Med. 2020, 39, 703–705. DOI: 10.1016/j.accpm.2020.10.006. DOI: https://doi.org/10.1016/j.accpm.2020.10.006
Kim, K.-D.; Hwang, I.; Ku, K. B.; Lee, S.; Kim, S.-J.; Kim, C. J. Microbiol. Biotechnol. 2020, 30, 1109–1115. DOI: 10.4014/jmb.2006.06006. DOI: https://doi.org/10.4014/jmb.2006.06006
Barton, M. I.; MacGowan, S. A.; Kutuzov, M. A.; Dushek, O.; Barton, G. J.; van der Merwe, P.A. Elife. 2021, 10. DOI: 10.7554/eLife.70658. DOI: https://doi.org/10.7554/eLife.70658
Li, C.; Tian, X.; Jia, X.; Wan, J.; Lu, L.; Jiang, S.; Lan, F.; Lu, Y.; Wu, Y.; Ying, T. Signal Transduct. Target. Ther. 2021, 6, 132. DOI: 10.1038/s41392-021-00536-0. DOI: https://doi.org/10.1038/s41392-021-00536-0
Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Science. 2005, 309, 1864–8. DOI: 10.1126/science.1116480. DOI: https://doi.org/10.1126/science.1116480
Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Science (80-.). 2020, eabb2762. DOI: 10.1126/science.abb2762. DOI: https://doi.org/10.1126/science.abb2762
Rui, L.; Haonan, L.; Wanyi, C. Biophys. Chem. 2020, 267, 106472. DOI: 10.1016/j.bpc.2020.106472. DOI: https://doi.org/10.1016/j.bpc.2020.106472
Vique‐Sánchez, J. L. Biointerface Res. Appl. Chem. 2021, 12, 5234–5265. DOI: 10.33263/BRIAC124.52345265. DOI: https://doi.org/10.33263/BRIAC124.52345265
de Oliveira, O. V.; Rocha, G. B.; Paluch, A. S.; Costa, L.T. J. Biomol. Struct. Dyn. 2021, 39, 3924–3933. DOI: 10.1080/07391102.2020.1772885. DOI: https://doi.org/10.1080/07391102.2020.1772885
Pirolli, D.; Righino, B.; De Rosa, M. C. Mol. Inform. 2021, 40, 2060080. DOI: 10.1002/minf.202060080. DOI: https://doi.org/10.1002/minf.202060080
Wang, L.; Wu, Y.; Yao, S.; Ge, H.; Zhu, Y.; Chen, K.; Chen, W.; Zhang, Y.; Zhu, W.; Wang, H.; et al. Acta Pharmacol. Sin. 2021. DOI: 10.1038/s41401-021-00735-z. DOI: https://doi.org/10.1038/s41401-021-00735-z
Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Cell. Mol. Immunol. 2020. DOI: 10.1038/s41423-020-0400-4. DOI: https://doi.org/10.1038/s41423-020-0400-4
Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. Nucleic Acids Res. 2018, 46, W296–W303. DOI: 10.1093/nar/gky427. DOI: https://doi.org/10.1093/nar/gky427
Swissmodel. https://swissmodel.expasy.org/, accessed in March 2022
RAMPAGE (RRID:SCR_017590). https://scicrunch.org/resolver/RRID:SCR_017590 , accessed in March 2022.
Brooks, B.R.; Brooks, C.L.; Mackerell, A.D.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. J. Comput. Chem. 2009, 30, 1545–1614. DOI: 10.1002/jcc.21287. DOI: https://doi.org/10.1002/jcc.21287
Halgren, T. A. J. Comput. Chem. 1996, 17, 490–519. DOI: 10.1002/(SICI)1096-987X(199604)17:5/6<490::AID-JCC1>3.0.CO;2-P. DOI: https://doi.org/10.1002/(SICI)1096-987X(199604)17:6<490::AID-JCC1>3.3.CO;2-V
ChemBridge Corp. https://chembridge.com/screening_libraries/#EXPRESSPick, accessed in January 2022
Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Adv. Drug Deliv. Rev. 2001, 46, 3–26. DOI: https://doi.org/10.1016/S0169-409X(00)00129-0
Thangapandian, S.; John, S.; Lee, Y.; Kim, S.; Lee, K.W. Int. J. Mol. Sci. 2011, 12, 9440–9462. DOI: 10.3390/ijms12129440. DOI: https://doi.org/10.3390/ijms12129440
Vique-Sánchez, J. L. Bioorganic Med. Chem. 2021, 33. DOI: 10.1016/j.bmc.2021.116040. DOI: https://doi.org/10.1016/j.bmc.2021.116040
Benítez-Cardoza, C. G.; Vique-Sánchez, J. L. Life Sci. 2020, 117970. DOI: 10.1016/j.lfs.2020.117970. DOI: https://doi.org/10.1016/j.lfs.2020.117970
Soga, S.; Shirai, H.; Kobori, M.; Hirayama, N. J. Chem. Inf. Model. 2007, 47, 400–406. DOI: 10.1021/ci6002202. DOI: https://doi.org/10.1021/ci6002202
Naïm, M.; Bhat, S.; Rankin, K.N.; Dennis, S.; Chowdhury, S.F.; Siddiqi, I.; Drabik, P.; Sulea, T.; Bayly, C.I.; Jakalian, A.; et al. J. Chem. Inf. Model. 2007, 47, 122–133. DOI: 10.1021/ci600406v. DOI: https://doi.org/10.1021/ci600406v
Labute, P. J. Comput. Chem. 2008, 29, 1693–8. DOI: 10.1002/jcc.20933. DOI: https://doi.org/10.1002/jcc.20933
Wadood, A.; Ghufran, M.; Hassan, S.F.; Khan, H.; Azam, S.S.; Rashid, U. Pharm. Biol. 2017, 55, 19–32. DOI: 10.1080/13880209.2016.1225778. DOI: https://doi.org/10.1080/13880209.2016.1225778
acdlabs. https://www.acdlabs.com/products/percepta/index.php, accessed in March 2022
PreADMET. https://preadmet.bmdrc.kr/toxicity/, accessed in March 2022
Grifoni, A.; Sidney, J.; Zhang, Y.; Scheuermann, R. H.; Peters, B.; Sette, A. Cell Host Microbe. 2020. DOI: 10.1016/j.chom.2020.03.002. DOI: https://doi.org/10.1016/j.chom.2020.03.002
Ton, A.-T.; Gentile, F.; Hsing, M.; Ban, F.; Cherkasov, A. Mol. Inform. 2020, minf.202000028. DOI: 10.1002/minf.202000028. DOI: https://doi.org/10.1002/minf.202000028
Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Science (80-.). 2020, eabb3405. DOI: 10.1126/science.abb3405. DOI: https://doi.org/10.1126/science.abb3405
Khelfaoui, H.; Harkati, D.; Saleh, B. A. J. Biomol. Struct. Dyn. 2020, 1–17. DOI: 10.1080/07391102.2020.1803967. DOI: https://doi.org/10.1080/07391102.2020.1803967
Focosi, D.; Maggi, F.; Franchini, M.; McConnell, S.; Casadevall, A. Int. J. Mol. Sci. 2021, 23, 29. DOI: 10.3390/ijms23010029. DOI: https://doi.org/10.3390/ijms23010029
Spinello, A.; Saltalamacchia, A.; Magistrato, A. J. Phys. Chem. Lett. 2020, 11, 4785–4790. DOI: 10.1021/acs.jpclett.0c01148. DOI: https://doi.org/10.1021/acs.jpclett.0c01148
Han, D. P.; Penn-Nicholson, A.; Cho, M. W. Virology. 2006, 350, 15–25. DOI: 10.1016/j.virol.2006.01.029. DOI: https://doi.org/10.1016/j.virol.2006.01.029
Vique-Sánchez, J. L.; Caro-Gómez, L. A.; Brieba, L. G.; Benítez-Cardoza, C. G. Parasitol. Int. 2020. DOI: 10.1016/j.parint.2020.102086. DOI: https://doi.org/10.1016/j.parint.2020.102086
Vique‐Sánchez, J. L.; Jiménez‐Pineda, A.; Benítez‐Cardoza, C. G. Arch. Pharm. (Weinheim). 2020. DOI: 10.1002/ardp.202000263. DOI: https://doi.org/10.1002/ardp.202000263
Benítez-Cardoza, C. G.; Fernández-Velasco, D.A.; Vique-Sánchez, J. L. ChemistrySelect. 2020. DOI: 10.1002/slct.201904632. DOI: https://doi.org/10.1002/slct.201904632
Arroyo-Verástegui, R.; Ortega-López, J.; Benítez-Cardoza, C.; Vique-Sánchez, J. L.; Brieba de castro, L. G.; Rojo-Domínguez, A.; García-Gutiérrez, P. El uso de 5,5´- [(4-nitrofenil)-metilen]bis(6-hidroxi-2-mercapto-3-metil-4(3H)-pirimidinonaTIM como tricomonicida. 2016.
ADMETlab. http://admet.scbdd.com/calcpre/index_sys/, accessed in March 2022.

Downloads
Additional Files
Published
Issue
Section
License
Copyright (c) 2022 Claudia Guadalupe Benítez-Cardoza, Jesús Néstor Ramirez-Torres, José Luis Vique-Sánchez

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.